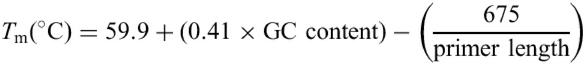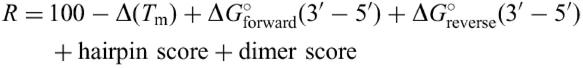Abstract
Primer Design Assistant (PDA) is a web interface primer design service combined with thermodynamic theory to evaluate the fitness of primers. It runs in a Linux–Apache–MySQL–PHP structure on a PC equipped with dual CPU (Intel Pentium III 1.4 GHz) and 512 Mb of RAM. A succinct user interface of PDA is accomplished by built-in parameters setting. Advanced options on 5′ GC content, 3′ GC content, dimer check and hairpin check are available. The option of covered region constrains the PCR product to cover a user-defined segment. PDA accepts single sequence query or multiple ones in FASTA format. It produces optimal and homogenous primer pairs that meet the need in experimental design with large-scaled PCR amplifications. Considering the system loading, the size of a submitted sequence is limited to 10 kb and the total sequence number in a query is limited to 20. The authors may be contacted regarding other requirements for primer design. The web application can be found at http://dbb.nhri.org.tw/primer/.
INTRODUCTION
PCR (polymerase chain reaction) is one of the most popular methods in biological and biomedical benchwork today. The novelty of PCR is that it uses the common chemical language among living things, the nucleic acid sequences, and increases it from a small amount into millions to billions. It opens a new age in genetic analysis on a molecular level (1,2). PCR is time-saving and sensitive, but the false positive signal in the detection is a serious problem. The most common causes of the artifact are the self-dimerization of primers and inadequate annealing of primers to an irrelevant template.
Primer design is fundamentally important in PCR-based detection methods. The general criteria for primers are very simple (3,4), yet it is difficult to choose good primers for a given template sequence. Not only are the calculating tasks heavy, a ranking mechanism for optimization is also very sophisticated. Therefore the computational aid on primer design is a critical issue in bioinformatics.
There are several web-based services or stand-alone software provided to the public for primer design, such as PRIDE (5), PRIMER MASTER (6), PRIMO (7), PrimeArray (8), Primer3 (4), Prime (9) and Web Primer (http://genome-www2.stanford.edu/cgi-bin/SGD/web-primer). Users can define the parameters listed in the menu of these tools and then get several pairs of primers for the target template sequence. However, most of them only take a single sequence query. Besides, the calculation of the primer annealing condition is often simplified in a short equation of melting temperature (Tm), regardless of the sequence content of the primer itself. As presented here, we provide a web interface primer design service, the Primer Design Assistant (PDA), combined with thermodynamic theory (10) to evaluate the fitness of primers. Multiple sequence query in a batch-wise mode is allowed. The advantage for the uniformity of primer annealing conditions would be helpful in experimental design with large-scaled PCR amplifications, such as probes prepared from phage clones preserved in a 96-well formatted plate, or any other applications that one may wish to amplify the target template(s) by different primer pairs at the same PCR run time.
IMPLEMENTATION
Pipeline of Primer Design Assistant (PDA)
The PDA service runs on a SMP (symmetric multi-processor) PC equipped with two CPU (Intel Pentium-III 1.4 GHz) and 512 Mb of RAM. We adopted the LAMP structure which is the abbreviation of Linux (Mandrake 9.0, the operating system)–Apache (version 1.3.27, the web server)–MySQL (version 4.0.10 gamma, the relational database)–PHP (version 4.3.0, the html embedded scripting language).
For each query sequence character string, a temporary database of substrings of user-defined length ‘product length’ is built. Forward and reverse primers are then picked from 5′ and 3′ terminals in a user defined ‘primer length’. Filtrating through the limits set in basic criteria and user-defined options, the survived primer pairs are sorted by a ranking mechanism discussed later.
Basic criteria for primer selection
Melting temperature
The primer annealing temperature (Ta) is determined by the melting temperature (Tm). The Tm of a given primer depends on several physiochemical factors. Primer Tm is calculated in the following equation (11).
In PDA default setting, the GC content of forward and reversed primers ranges from 30 to 70%. Primers with Tm lower than 50°C are excluded. The acceptable difference of Tm values in a primer pair is within 5°C.
Sequence complexity
Low complexity on primer will reduce discriminating power on the sequence content. In PDA, we exclude any four or more continual nucleotides, such as AAAA, TTTT, CCCC or GGGG for both forward and reversed primers. Continuous di-nucleotide repeats, such as ‘ATATAT’, are also avoided.
3′ C/G clamp
Nucleotide residue ‘C’ or ‘G’ forms a stronger pairing structure in the duplex DNA strands. The stable 3′ end in primer template complex will improve the polymerase efficiency (12). In PDA, 3′ C/G clamp is set as default to constrain the selected forward primer and reverse primer to end up in C or G.
5′ GC content and 3′ GC content
The GC content in the first six nucleotide residues (5′ GC content) and in the last six nucleotide residues (3′ GC content) is evaluated in order to enhance the priming specificity. In PDA options, we define that the 5′ GC content is not less than 50% and the 3′ GC content ranges from 30 to 60%.
Computation of secondary structure formation
Primer dimerization
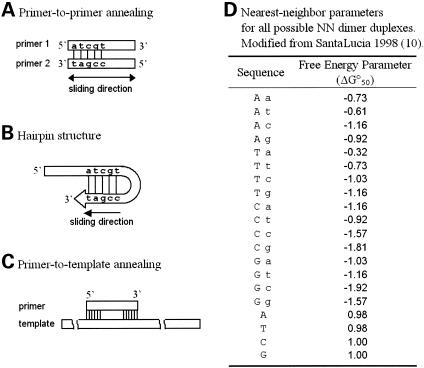
Two types of primer-dimer, self-dimer and cross-dimer, may occur in a PCR reaction. The stability of each pairing condition is calculated by a Nearest-Neighbor model (13,14) (Fig. 1A). All possible pairing conditions are examined by stepwise sliding one primer against another forward or backward until the overlapping length is down to 4. The sum of scores from all pairing conditions is defined as the dimer score for a given primer pair.
Figure 1.
Calculation of the stability of DNA duplexes. The stability of DNA duplex structure in primer-to-primer annealing (primer-dimer, A), hairpin loop structure (B) and primer-to-template annealing (C) is estimated by nearest-neighbor dimer sequence in the pairing segment with NN parameters (D) as follows. When sequence 1 [5′- atcgt -3′] aligns to sequence 2 [3′- tagcc- 5′], the first base of the first sequence (a) matches to (t) in sequence 2, and follows with three more Watson–Crick pairs. The fifth base mismatches. The NN propagation energy of the continuing base pairs (at), (tc), (cg) and the mismatched base (t) in primer 1 are summed up: (−0.61)+(−1.03)+(−1.81)+(0.98)=−2.47.
Hairpin loop
The potency of hairpin loop formation is estimated in a similar way. We ‘bend’ the 3′ end of a given primer backward in length by four and slide stepwise against itself. The hairpin structure stability is calculated in all temporary sliding matches for each primer by a Nearest-Neighbor model (Fig. 1B). The sum of scores for a primer pair is defined as hairpin score.
Ranking mechanism
To avoid the mis-priming amplification, the 5′ end of the primer is expected to anneal to target templates more stable than the 3′ end (15). Again we apply a Nearest-Neighbor model to estimate the stability of the primer-to-template pairing (Fig. 1C). The difference on free energy (ΔG°) between the 3′ end (the last six bases) and 5′ end (the first six bases) is calculated by the equation: ΔG°(3′−5′)=ΔG°3′−ΔG°5′.
Finally, the primer pairs passing through the limitation listed above are sorted by ranking score (R):
PDA INTERFACE
Input interface
The screenshot of the PDA input form is shown in Figure 2. Users may paste the target DNA into the sequence input form in both flat text ASCII and FASTA format. Multiple sequences submission in FASTA format is allowed. All numbers, spaces and line breaks will be trimmed out automatically. PDA is case-insensitive. Characters other than the typical ‘A T C G’ will be replaced by ‘N’.
Figure 2.
Web interface of Primer Design Assistant.
The primer length ranges from 18 to 25 nt and the default setting is 19. The expected product size ranges from 150 to 600 bp and the default setting is 150. A succinct user interface of PDA is accomplished by a built-in parameters setting including Tm range, GC content, 3′ GC clamp, sequence complexity check and difference on Tm. It meets most investigators' needs in primer design, without being confusing with numerous options to choose from. Four advanced parameters including dimer check, hairpin check, 5′ GC content check and 3′ GC content check are optional. In our experience, primer design on some sequences may fail to pass through the 5′ GC content and 3′ GC content check; but in most cases, primers without these checks may work as well in the laboratory. The option of covered region constrains the PCR product to cover a user-defined segment by entering the start and end position of the submitted sequence.
Output result
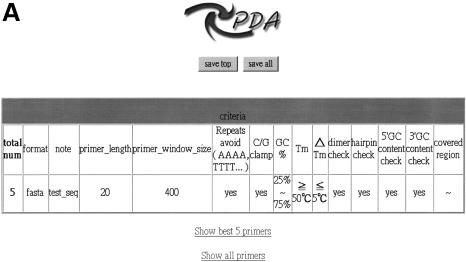
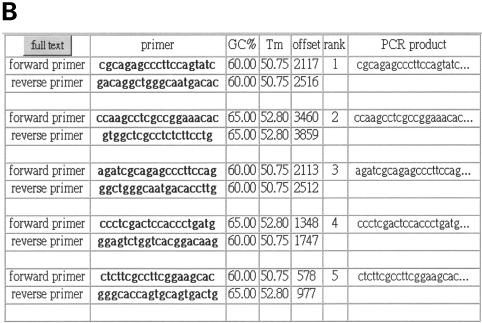
When a query is submitted, PDA first returns a brief set of parameters (Fig. 3A). Users may follow the hyperlink ‘show best five primers’ or ‘show all primers’ (Fig. 3B) to read the result in a web browser. Furthermore, users may download the returned results in a text format with tab separation or open it in a spreadsheet application such as MS-Excel. The best primer pair for each query sequence will be retrieved in ‘save top’ option and all matched primer pairs in ‘save all’.
Figure 3.
The output of Primer Design Assistant. (A) The summary of parameters setting on a query. (B) The example for output results following the link ‘Show best 5 primers’ in (A).
SYSTEM PERFORMANCE
In a collaborative microarray project, we attempted to generate primer pairs to amplify EST clones stored in a 96-well formatted plate. Generally, clones were amplified with primers on vector arms. The amplified products vary in length, depending on the size of the insertion. Consequently, the inconsistency of hybridization efficiency may lead to a serious problem in quantification of spot intensity. In the batch-wise mode, we selected primer pairs for these EST clones and restricted the length of the amplified product. Except for the empty wells or misplaced clones, the percentage of successfully amplified clones reached 99%.
The system performance of PDA was optimized and tested on an input sequence (AP002939: 1–10 000) with 10 000 bases and a file containing five sequences (near 13 000 bp) in FASTA format (these datasets can be found on the PDA web site). In the case when the advanced options were set ‘off’, it took 4.12 (a single sequence query) and 20.21 s (a five sequence query) to complete the job. When the options were set ‘on’, 4.05 and 12.63 s were spent passing through the checking mechanism, respectively. Due to strict checking mechanisms by advanced options, only qualified candidates should be calculated. Using PDA with advanced options will get fewer primer pairs and decrease the time of calculation, otherwise.
Considering the system loading, the size of submitted sequence is limited to 10 kb and the total sequence number in a query is limited to 20. Other requirements for primer design, such as high-throughput experiments, may be forwarded to the authors.
Acknowledgments
ACKNOWLEDGEMENTS
We are deeply grateful to Dr J.L. Juang, the assistant investigator of NHRI, for his help in evaluating the performance of the primer pairs designed by PDA, and our colleague Dr Yen-Ching Chen, for her comments on the article. Our thanks also go to the anonymous referees who read the manuscript carefully and gave us many valuable suggestions.
REFERENCES
- 1.Glennon M. and Cormican,M. (2001) Detection and diagnosis of mycobacterial pathogens using PCR. Expert. Rev. Mol. Diagn., 1, 163–174. [DOI] [PubMed] [Google Scholar]
- 2.Jain K.K. (2002) Current trends in molecular diagnostics. Med. Device Technol., 13, 14–18. [PubMed] [Google Scholar]
- 3.Robertson J.M. and Walsh-Weller,J. (1998) An introduction to PCR primer design and optimization of amplification reactions. Methods Mol. Biol., 98, 121–154. [DOI] [PubMed] [Google Scholar]
- 4.Rozen S. and Skaletsky,H. (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol., 132, 365–386. [DOI] [PubMed] [Google Scholar]
- 5.Haas S., Vingron,M., Poustka,A. and Wiemann,S. (1998) Primer design for large scale sequencing. Nucleic Acids Res., 26, 3006–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Proutski V. and Holmes,E.C. (1996) Primer Master: a new program for the design and analysis of PCR primers. Comput. Appl. Biosci., 12, 253–255. [DOI] [PubMed] [Google Scholar]
- 7.Li P., Kupfer,K.C., Davies,C.J., Burbee,D., Evans,G.A. and Garner,H.R. (1997) PRIMO: a primer design program that applies base quality statistics for automated large-scale DNA sequencing. Genomics, 40, 476–485. [DOI] [PubMed] [Google Scholar]
- 8.Raddatz G., Dehio,M., Meyer,T.F. and Dehio,C. (2001) PrimeArray: genome-scale primer design for DNA-microarray construction. Bioinformatics, 17, 98–99. [DOI] [PubMed] [Google Scholar]
- 9.Eberhardt N.L. (1992) A shell program for the design of PCR primers using genetics computer group (GCG) software (7.1) on VAX/VMS systems. Biotechniques, 13, 914–917. [PubMed] [Google Scholar]
- 10.SantaLucia J. Jr. (1998) A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA, 95, 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freier S.M., Kierzek,R., Jaeger,J.A., Sugimoto,N., Caruthers,M.H., Neilson,T. and Turner,D.H. (1986) Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl Acad. Sci. USA, 83, 9373–9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buck G.A., Fox,J.W., Gunthorpe,M., Hager,K.M., Naeve,C.W., Pon,R.T., Adams,P.S. and Rush,J. (1999) Design strategies and performance of custom DNA sequencing primers. Biotechniques, 27, 528–536. [DOI] [PubMed] [Google Scholar]
- 13.SantaLucia J. Jr., Allawi,H.T. and Seneviratne,P.A. (1996) Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry, 35, 3555–3562. [DOI] [PubMed] [Google Scholar]
- 14.Sugimoto N., Nakano,S., Yoneyama,M. and Honda,K. (1996) Improved thermodynamic parameters and helix initiation factor to predict stability of DNA duplexes. Nucleic Acids Res., 24, 4501–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breslauer K.J., Frank,R., Blocker,H. and Marky,L.A. (1986) Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA, 83, 3746–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]