Abstract
Splicing is a biological phenomenon that removes the non-coding sequence from the transcripts to produce a mature transcript suitable for translation. To study this phenomenon, information on the intron–exon arrangement of a gene is essential, usually obtained by aligning mRNA/EST sequences to their cognate genomic sequences. MGAlign is a novel, rapid, memory efficient and practical method for aligning mRNA/EST and genome sequences. We present here a freely available web service, MGAlignIt (http://origin.bic.nus.edu.sg/mgalign/mgalignit), based on MGAlign. Besides the alignment itself, this web service allows users to effectively visualize the alignment in a graphical manner and to perform limited analysis on the alignment output. The server also permits the alignment to be saved in several forms, both graphical and text, suitable for further processing and analysis by other programs.
INTRODUCTION
The completion of a draft human genome (1,2) in February 2001 revealed that the human genome has only ∼30 000–40 000 genes, compared to the initial estimate of 100 000 genes, to account for the complexity in the species. Drafts of numerous other genomes (3–8) have been completed and once again, it is observed that the number of genes do not increase greatly with the complexity of the organism. However, the number of proteins in the organism is far in excess of the number of genes, as there exist numerous cellular mechanisms that lead to multiple gene products from a single gene. Alternative splicing, the differential joining of exons, is one of these mechanisms and perhaps the most important one (9–11).
To understand and study splicing and its implications, we need to determine the intron–exon arrangement in a gene, typically unraveled by performing a sequence alignment of the mRNA sequence of the gene with its cognate genomic sequence (12). A database of human and mouse genes showing alternate exon arrangements is an example of such a study (Alternate Exon Database, http://www.ebi.ac.uk/asd/altextron/access.html). This approach produces a global alignment consisting of several local alignments, with each local alignment being a single exon. The usefulness of this approach in obtaining intron–exon arrangement information is enhanced by the fact that complete genomic sequences of several organisms are now available, thus providing part of the data for such alignments. Other than genome sequencing projects, there are several projects seeking to generate full-length cDNA sequences (13,14) of organisms under way. Besides full-length cDNA sequences, there exist a huge amount of ESTs (expressed sequence tags) in public databases like dbEST (15). The availability of these resources means that obtaining intron–exon arrangement information of genes from sequence alignments of mRNA/EST sequences to genomic sequences is very practical nowadays.
MGAlign (Lee,B.T.K., Tan,T.W. and Ranganathan,S., unpublished results) is a new method that aligns mRNA/EST sequences to genomic sequences using a rapid heuristic method. A web interface at http://origin.bic.nus.edu.sg/mgalign/mgalignit.html that utilizes MGAlign as the alignment technique is available for users to easily visualize the intron–exon arrangements and to perform limited analysis on the alignments. In this paper, we will describe the web service, MGAlignIt, as an alignment service.
OVERVIEW OF THE WEB SERVICE
The MGAlignIt web service provides a dynamic means of aligning pairs of mRNA/EST and genomic sequences at the same website, based on MGAlign, a method that rapidly aligns an mRNA/EST sequence to its cognate genomic sequence using a heuristic approach. The software is available in binary form for several platforms (Microsoft Windows, Sun Solaris, Linux, Mac OS X and SGI IRIX) at http://origin.bic.nus.edu.sg/mgalign. The web server aims to provide the global research community with a free alignment portal for mRNA/EST and genomic sequences, using MGAlign, with output options enabling simple yet intuitive visualization of the intron–exon arrangement graphically, a rapid means of determining the effects of alternative splicing on the alignment, the ability to perform limited analysis on the alignment and the facility to save the alignment in various formats for integration with other tools.
MGAlign fulfils these collective aims using a three-step process. The first step tackles the alignment itself, by basically aligning the mRNA/EST and genomic sequences. This is followed by a visualization step where the user can visualize and perform limited analysis on the alignment. Lastly, the user is able to request outputs of the alignment in various forms, for local analysis, publication and presentation.
ALIGNMENT
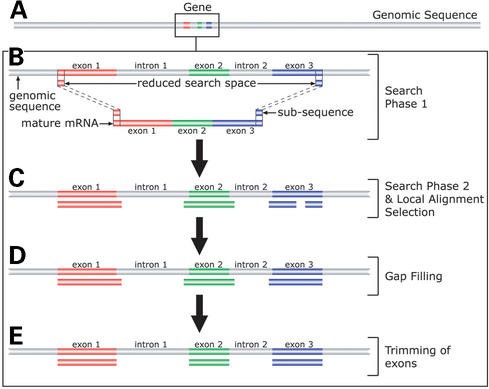
A schematic diagram of the algorithm used by MGAlign is shown in Figure 1. The distinguishing feature of the algorithm is in the heuristics used to reduce the search space. MGAlign achieves this reduction in search space by using two search phases instead of one. The first search phase aims to locate for a pair of matches on the genomic sequence such that the rest of the alignment lies between this pair of matches as shown in Figure 1B. Thus the algorithm has only to search within this segment of genomic sequence bounded by the pair of matches for the rest of the alignment. In the event that more than one pair of matches is found, the process given in Figure 1 is repeated for each pair of matches and the one with the best score reported. After the pair of matches is found, sub-sequences of the mRNA sequence is then used to locate for local alignments within the segment of genomic sequence bounded by the pair of matches. There is a possibility that the same sub-sequence will result in more than one local alignment. Therefore the algorithm has to select a subset of the local alignments that maximizes the amount of aligned mRNA sequence as shown in Figure 1C. The next two steps involve filling in of gaps using shorter sub-sequences and trimming of overlapping exons using information on splice site motifs as illustrated in Figure 1D and E. More details of the algorithm are described elsewhere (Lee,B.T.K., Tan,T.W. and Ranganathan,S., unpublished results), with benchmarking results indicating that MGAlign is more accurate and faster than sim4 and Spidey, with only limited memory requirements (available from http://origin.bic.nus.edu.sg/mgalign/comparison.html).
Figure 1.
Schematic diagram of MGAlign's approach. (A) The box represents the region occupied by the mRNA/EST sequence on the genomic sequence (grey bar). (B) Search Phase 1 locates matches on the genomic sequence using short sub-sequences from the ends of the mRNA sequence, to reduce the alignment space. (C) Search Phase 2 locates regions of local alignment use by searching within the reduced search space. (D) Alignment gaps are filled by searching with smaller sub-sequences. (E) Lastly overlaps between the exons are trimmed based in splice site motifs.
VISUALIZATION
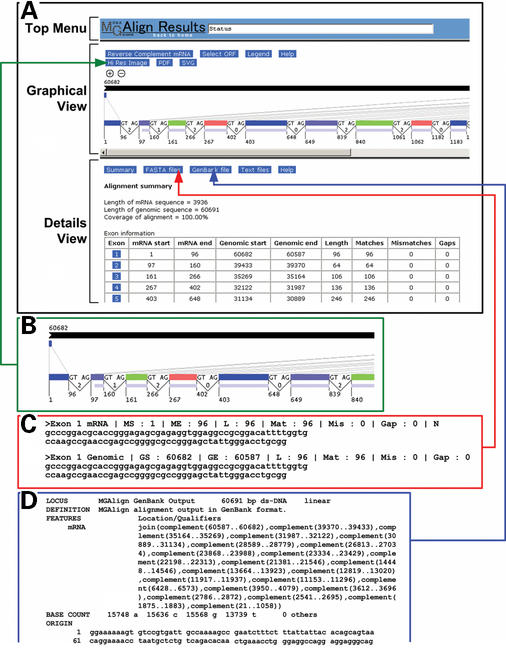
Once the alignment is finished, the output of the algorithm is parsed into a web page (Fig. 2) consisting of three frames to visualize and analyze the alignment. The top frame entitled ‘Top Menu’ includes a status bar that provides feedback and instructions to users as well as a link to bring the users back to the homepage. The middle frame named ‘Graphical View’ provides a graphical representation of the alignment [a Portable Network Graphics (PNG) image], which can be enlarged or reduced in size via the zoom in/out buttons for better display. This is especially useful for users as it provides an overall view of the alignment, giving them a sense of the length and distribution of the exons and introns. The genome sequence is represented by a black bar, followed by the mRNA/EST sequence, represented in two ways. The top series of colored bars (each bar corresponding to an individual exon) have their dimensions and positions scaled relative to the length of the genomic sequence. This gives the users a feel of the size and distribution of each exon as compared to the genomic sequence. The use of alternating colors helps in differentiating exons, which could be a problem when they are closely packed. The bottom series of colored bars have their sizes scaled to the length of the mRNA/EST sequence. This provides users a means of estimating the relative size and location of each exon and to aid in correlating the exons from the top series to the bottom series, connected by grey lines. The splice site motifs, intron phases and the start and end positions of the exons are indicated on the bottom series of colored bars. This allows users to determine at a glance if there are any non-canonical splice sites and the effects of any alternative splicing. For example, Figure 2B shows the 5′ portion of an alignment, where a frame shift in the coding sequence will result if exon 2 is spliced out, since exon 2 is flanked by introns of different phases. However, if exon 5 were to be spliced out, there would not be any frame shift as the introns bordering exon 5 are in phase. To further aid in determining the effects of alternative splicing, the longest open reading frame (ORF) of the mRNA sequence is drawn below the second series of colored bars as a light blue colored bar, allowing easy identification of the coding region and rapid determination of the effects of any frame shifts. For example, insertion of a frame shift-inducing exon between exons 1 and 2 would not affect the coding sequence as the start of the coding sequence lies within exon 2. As the mRNA/EST sequence could potentially have several ORFs, there is an option to select which ORF is to be used in the image. The amino acid sequence corresponding to each ORF is also linked to the BLAST (16) search page at NCBI (National Center for Biotechnology Information) for fast protein identification. The image of the alignment is itself an intuitive navigational tool. Users can get detailed information about specific parts of the alignment by clicking on elements in the image. Selecting an exon by clicking on it, provides detailed information about that exon in the bottom frame.
Figure 2.
Results from the MGAlignIt web server. (A) The main results page is divided into three frames entitled ‘Top Menu’, ‘Graphical View’ and ‘Details View’. (B) Clicking the ‘Hi Res Image’ button leads to the high-resolution image, part of which is shown here. (C) Alignments of individual exons, introns and ORFs can be obtained by clicking on the ‘FASTA files’ button. The FASTA alignment for exon 1 is shown here. (D) A GenBank formatted record of the alignment is presented by clicking the ‘GenBank file’ button.
The bottom frame entitled ‘Details View’ provides additional information about specific portions of the alignment, such as the pair-wise alignment view of an exon with its cognate genomic sequence. The information to be displayed in this frame is controlled by selecting specific elements in the image found in the ‘Graphical View’ and the buttons found in the ‘Details View’. The interface is designed such that users can focus on the information they require. An instance of this flexible display option would be to view the alignment near a specific splice site. This is achieved by selecting the specific intron in the image in the ‘Graphical View’ and then clicking on the button entitled ‘Splice site details’ in the ‘Details View’. The desired information is then displayed in the ‘Details View’. Users can also do limited analysis within this frame. One such analysis is searching for patterns in both exonic and intronic sequences. This is especially useful for determining the existence of specific signals like branch sites in intronic sequences or exonic splicing enhancers in exonic sequences. Users can opt to use their own patterns or they may select one of the predefined ones available from the MGAlignIt website. The syntax followed is the one prescribed by PROSITE (17) allowing rapid and flexible searches. Besides pattern searches, exonic sequences and ORFs are linked to the NCBI's BLAST search page, permitting immediate access to homolog detection, by searching the GenBank (18) database.
Links to online help is provided on every MGAlignIt page. Selecting these links provide users with context specific help. In addition, there is an online tutorial on using the web services and as well as comprehensive help pages for all aspects of the output. An email helpline is also available for MGAlignIt users.
OUTPUT OPTIONS
Besides a simple visual representation of the alignment on the web page, the web service also provides many forms of output that users can save on their local computers. The graphical view of the alignment can be saved in three different formats. The first of these formats is a high resolution PNG raster image of the alignment, which appears in a new window when users click on ‘Hi Res Image’ in the ‘Graphical View’ frame. This provides users with a commonly used image format that can be incorporated into many applications. To facilitate scaling the image size and high quality printing, two additional formats are available. In the PDF (Portable Document Format) format, the alignment is shown as an image in an A4-sized PDF file, which users can save onto their computers. This format is compact and excellent for printing, as the image has already been resized to fill the paper and can be viewed and printed from any computer platform via a PDF viewer. The PDF file may also be imported and edited using common graphic applications such as Adobe Illustrator, specially for adding custom annotations to the alignment. The last graphical format is SVG (Scalable Vector Graphics), which is also a vector-based image format recommended as the standard by the W3C (the World Wide Web Consortium) that provides similar advantages as the PDF format. Applications such as Adobe Illustrator can be used to edit and annotate SVG images.
Other than graphical representations, the alignment can also be saved in two commonly used sequence formats, FASTA and GenBank. Users can obtain FASTA files of individual exons or introns as well as composite files containing all introns or all exons. Useful information about the alignment is stored in the header line of the FASTA files in a format that is easy for computers to parse. Each exon is stored as two FASTA sequences, one being the exonic portion of the alignment and the other being the genomic portion of the alignment as seen in Figure 2C. The FASTA format is a widely used format and making the alignment available in the FASTA format means that many other software can read the alignment generated. This permits incorporation of the alignment into other programs for further analysis.
The GenBank formatted record does not have the pairwise alignment information of each exon but it does allow for annotation of the genomic sequence in the Feature Table portion of the record. The alignment is annotated as an mRNA feature in the Feature Table as shown in Figure 2D. This facilitates the alignment to be used in other programs that read GenBank formatted records such as Artemis (19). Artemis is a standalone DNA sequence visualization and annotation tool.
The users can also save summaries of the exon, intron and ORF information. These files are provided as comma delimited text files, such that they can be viewed and formatted by several spreadsheet programs such as Microsoft Excel. This text output format is a very useful companion to the graphical output as the graphical output does not provide the full details of the alignment.
CONCLUSION
The MGAlignIt web service provides a platform for generating alignments of mRNA/EST sequences to their cognate genomic sequences in a freely accessible manner. In addition to providing a means for alignment, the web service makes possible easy visualization of the alignment, allowing users to quickly determine the effects of any changes in the splicing pattern. Furthermore, the web service allows further analysis such as BLAST searches and pattern searching. Lastly users can obtain soft copies of the alignment in several formats, including high resolution graphics, for further analysis, publication and printing.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank our colleagues at the Department of Biochemistry, National University of Singapore for their helpful comments and discussions. B.T.K.L. would also like to thank the National University of Singapore for the award of an Agency for Science, Technology and Research, Singapore (ASTAR) scholarship.
REFERENCES
- 1.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 2.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 3.Blattner F.R., Plunkett,G.,III, Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 4.Cherry J.M., Ball,C., Weng,S., Juvik,G., Schmidt,R., Adler,C., Dunn,B., Dwight,S., Riles,L., Mortimer,R.K. et al. (1997) Genetic and physical maps of Saccharomyces cerevisiae. Nature, 387, 67–73. [PMC free article] [PubMed] [Google Scholar]
- 5.Holt R.A., Subramanian,G.M., Halpern,A., Sutton,G.G., Charlab,R., Nusskern,D.R., Wincker,P., Clark,A.G., Ribeiro,J.M., Wides,R. et al. (2002) The genome sequence of the malaria mosquito Anopheles gambiae. Science, 298, 129–149. [DOI] [PubMed] [Google Scholar]
- 6.The Arabidopsis Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 7.The C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. The C. elegans Sequencing Consortium. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 8.Wood V., Gwilliam,R., Rajandream,M.A., Lyne,M., Lyne,R., Stewart,A., Sgouros,J., Peat,N., Hayles,J., Baker,S. et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature, 415, 871–880. [DOI] [PubMed] [Google Scholar]
- 9.Maniatis T. and Tasic,B. (2002) Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature, 418, 236–243. [DOI] [PubMed] [Google Scholar]
- 10.Black D.L. (2000) Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell, 103, 367–370. [DOI] [PubMed] [Google Scholar]
- 11.Graveley B.R. (2001) Alternative splicing: increasing diversity in the proteomic world. Trends Genet., 17, 100–107. [DOI] [PubMed] [Google Scholar]
- 12.Clark F. and Thanaraj,T.A. (2002) Categorization and characterization of transcript-confirmed constitutively and alternatively spliced introns and exons from human. Hum. Mol. Genet., 11. 451–464. [DOI] [PubMed] [Google Scholar]
- 13.Strausberg R.L., Feingold,E.A., Klausner,R.D. and Collins,F.S. (1999) The mammalian gene collection. Science, 286, 455–457. [DOI] [PubMed] [Google Scholar]
- 14.Okazaki Y., Furuno,M., Kasukawa,T., Adachi,J., Bono,H., Kondo,S., Nikaido,I., Osato,N., Saito,R., Suzuki,H. et al. (2002) Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature, 420, 563–573. [DOI] [PubMed] [Google Scholar]
- 15.Boguski M.S., Lowe,T.M. and Tolstoshev,C.M. (1993) dbEST—database for ‘expressed sequence tags’. Nature Genet., 4, 332–333. [DOI] [PubMed] [Google Scholar]
- 16.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 17.Falquet L., Pagni,M., Bucher,P., Hulo,N., Sigrist,C.J., Hofmann,K. and Bairoch,A. (2002) The PROSITE database, its status in 2002. Nucleic Acids Res., 30, 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J. and Wheeler,D.L. (2003) GenBank. Nucleic Acids Res., 31, 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rutherford K., Parkhill,J., Crook,J., Horsnell,T., Rice,P., Rajandream,M.A. and Barrell,B. (2000) Artemis: sequence visualization and annotation. Bioinformatics, 16, 944–945. [DOI] [PubMed] [Google Scholar]