Abstract
WEB-THERMODYN analyzes DNA sequences and computes the DNA helical stability, i.e. the free energy required to unwind and separate the strands of the double helix. A helical stability profile across a selected DNA region or the entire sequence is generated by sliding-window analysis. WEB-THERMODYN can predict sites of low helical stability present at regulatory regions for transcription and replication and can be used to test the influence of mutations. The program can be accessed at: http://wings.buffalo.edu/gsa/dna/dk/WEBTHERMODYN/.
INTRODUCTION
The stability of the double helix varies across a DNA molecule. Regulatory regions for replication and transcription sometimes have a lower helical stability than gene coding regions in prokaryotic and eukaryotic DNA. Localized opening of the double helix occurs at certain regulatory regions in supercoiled DNA at physiological temperature (1–5). Previously our laboratory developed a PC computer program, THERMODYN, to predict regions of low helical stability in DNA and to assess their functional contribution (6). The program calculates the helical stability from the DNA sequence and the known thermodynamic properties of the component nearest-neighbor nucleotides (7,8). DNA helical stability is defined as the free energy required for unwinding and separating the strands of the double helix. Experimentally, the low helical stability of regulatory regions has been detected by hypersensitivity to single-strand specific nucleases or chemical modification and by stable unwinding of DNA topoisomers seen after 2D gel electrophoresis (2,5; 9 and references therein). THERMODYN analysis correctly predicted the experimentally determined sites of low helical stability in several replication origins and gene terminator and promoter regions as well as the hierarchy of those sites in plasmids (10). Analysis of mutations in a yeast replication origin (ARS307/C2G1 ARS) indicated that the low helical stability region contains a DNA unwinding element (DUE) (6), a cis-acting regulatory element whose intrinsic helical instability is important for its biological function (4).
Here we describe WEB-THERMODYN, a platform independent version of the THERMODYN program with a user-friendly browser interface and several other advantages. WEB-THERMODYN can analyze large DNA sequences and create a graphical output of the helical stability profile. The program automates localization of low helical stability regions and links the regions to a display of the corresponding DNA sequences. Below we present an example of the WEB-THERMODYN output as well as information for the user concerning input, output and the program. Finally, the variety of replication origins and gene regulatory regions known to have low helical stability in vitro as well as the use of WEB-THERMODYN for predicting such regulatory regions and testing the influence of mutations are discussed.
RESULTS
Example of a helical stability profile with links to the DNA sequence
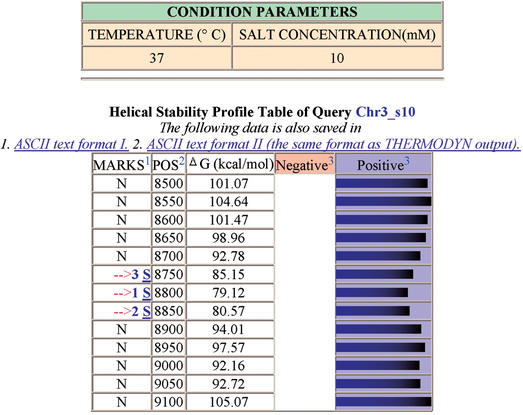
A helical stability profile of a yeast chromosome region containing two gene coding regions that flank a replication origin is illustrated at the bottom of an example output page (Fig. 1). The free energy minima map at the replication origin (ARS307) between the genes. Each of the free energy minima (Fig. 1, MARKS 1 S, 2 S and 3 S) is linked to a second output page (not shown) displaying the DNA sequence of the 100 bp window analyzed. In this example, the sequences corresponding to free energy minima 1 and 2 contain the ARS307 DUE region that is hypersensitive to a single-strand-specific nuclease and whose low helical stability is important for replication origin function (6).
Figure 1.
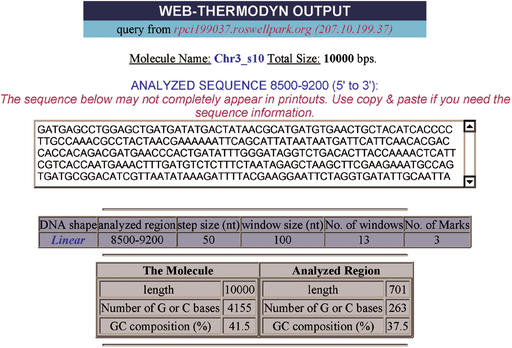
WEB-THERMODYN output page (see text). Analysis of an S.cerevisiae chromosome III segment (positions 100001 to 110000) in a region containing a replication origin (ARS307) and two gene coding regions. Free energy minima at marks 1 and 2 occur at sequence positions that overlap the replication origin in the DUE region (6). Gene coding regions with elevated free energies flank the positions of the energy minima at the replication origin.
WEB-THERMODYN input
Input values include the temperature (°C) and salt concentration (mM). Default values are supplied, and these are based on experimental conditions used to identify regions of low helical stability in plasmids (2). The users selects the DNA molecule shape, i.e. linear or circular, and enters the molecule name. The DNA sequence can be pasted into the browser window (maximum sequence size=30 kb) or typed. Alternatively, the sequence (≤40 kb) can be uploaded from a computer file and imported in various formats (ASCII/TXT, Genbank HTML/TXT, FASTA TXT, FASTA HTML). A, G, T and C are the only acceptable characters (no N's) and numbers are ignored.
The user selects to analyze either the sequence of the whole DNA molecule or a specific region. In the latter case, the user enters the start and end positions to define the region. Default values for window size (100 bp) and step size (50 bp) are supplied. The 100 bp window size approximates the length of low helical stability regions identified experimentally by mapping single-strand-specific nuclease nicks at nucleotide-level resolution (2). Either smaller or larger windows and step sizes may be appropriate for particular purposes. The number of energy minima markers (default=1) can be adjusted to highlight the lower free energy values in the output and to display the associated DNA sequences (see below). The default maximum computing time is set to 30 s, but this time may be increased to accommodate larger sequences and more steps, markers or windows.
WEB-THERMODYN output
An example output page for the analysis of a DNA molecule is shown (Fig. 1). The output displays the name of the DNA molecule, its total size and the analyzed sequence, if less than 30 kb. The parameters selected, including DNA shape, analyzed region, step size, window size and number of energy minima marks are displayed in a table. Included is the number of windows actually analyzed, based on the input parameters.
A table comparing the whole molecule and the analyzed region with respect to length, number of G or C bases and % G+C composition is displayed. Finally, a table of the condition parameters, i.e. temperature and salt concentration, is shown.
A helical stability profile table for the DNA molecule and a bar graph are displayed at the bottom of the output page (Fig. 1). In the first column of the table under ‘MARKS’, an N indicates that no energy minima marker is present. Where present, the energy minima marker (>) is displayed with its rank (1=lowest energy, 2=next lowest energy, etc.) and a sequence link, S (see below). The second column ‘POS’ shows the position of the first nucleotide in each window, and the third column ‘ΔG’ shows the free energy value for each window. The blue bars in the column under ‘Positive’ comprise a horizontal bar graph depicting the positive free energy values. The longer the bar, the greater the free energy. The shorter bars indicate the lower free energy values and predict the lower helical stability regions.
Clicking on a sequence link, S, next to an energy minimum marker displays another output page. That page allows the user to view the specific DNA sequence corresponding to the selected energy minimum.
Links to two types of ASCII text files are shown above the graphical output. One type indexes the position according to the first nucleotide in each sequence window, corresponding to the WEB-THERMODYN output. The other type indexes the position according to the central nucleotide in the sequence window, in the style of THERMODYN. Either file type may serve as the input for the user's graphical analysis software, but which type was used should be specified.
Sequences >30 kb are not displayed in the browser window on the output page, but are saved to a file for which a link is included. When the helical stability profile table is too large to display at once, the table is automatically subdivided into multiple files, and a list of links to those files is provided.
The program
The WEB-THERMODYN program was written in Practical Extraction and Report Language (PERL) and HyperText Markup Language (HTML) and uses the Common Gateway Interface (CGI) for input and output to a web browser. The free energy change calculated is that for the local separation of DNA strands in the context of a larger molecule that remains double helical. This process is independent of DNA concentration and thus the equations differ from those used for determining conventional DNA melting parameters. A description of the nearest-neighbor algorithm and the equations used to calculate DNA helical stability are provided at http://wings.buffalo.edu/gsa/dna/dk/WEBTHERMODYN/README.html.
DISCUSSION
WEB-THERMODYN permits facile profiling of DNA helical stability across a DNA sequence. At regulatory regions for replication and transcription, the low helical stability predicted by this and the related THERMODYN program has been experimentally validated by the hypersensitivity to a single-strand-specific nuclease in vitro (2,6,10). Low DNA helical stability in vitro is a physical property shared by replication origins from different species as well as from certain viruses and phage (3,4,11,12). Mutational analysis has indicated that a particular low helical stability sequence in the origin contains a DUE important for replication function (3,4,8,12). The DUE identified by mutational analysis corresponds to the DNA sequence that first unwinds during the intiation of replication in vitro (12–14). Matrix attachment regions involved in gene regulation also exhibit a low helical stability that is important for biological function (5,15–17). Recently, helical stability analysis using WEB-THERMODYN successfully predicted the locations of matrix attachment regions in human DNA (18). The ability of WEB-THERMODYN to analyse large DNA sequences and automate the search for free energy minima will facilitate identification of low helical stability regions associated with known and potential regulatory regions in genomic DNA. Finally, the use of WEB-THERMODYN to test the effects of mutations on helical stability using established approaches (6,8,19) will help identify the contributions that DNA helical stability makes to biochemical and biological functions.
Acknowledgments
ACKNOWLEDGEMENT
This research was supported by a grant from the National Institutes of Health (GM30614).
REFERENCES
- 1.Sheflin L.G. and Kowalski,D. (1985) Altered DNA conformations detected by mung bean nuclease occur in promoter and terminator regions of supercoiled pBR322 DNA. Nucleic Acids Res., 13, 6137–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kowalski D., Natale,D.A. and Eddy,M.J. (1988) Stable DNA unwinding, not ‘breathing’, accounts for single-strand-specific nuclease hypersensitivity of specific A+T-rich sequences. Proc. Natl Acad. Sci. USA, 85, 9464–9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Umek R.M. and Kowalski,D. (1988) The ease of DNA unwinding as a determinant of initiation at yeast replication origins. Cell, 52, 559–567. [DOI] [PubMed] [Google Scholar]
- 4.Kowalski D. and Eddy,M.J. (1989) The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J., 8, 4335–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohwi-Shigematsu T. and Kohwi,Y. (1990) Torsional stress stabilizes extended base unpairing in suppressor sites flanking immunoglobulin heavy chain enhancer. Biochemistry, 29, 9551–9560. [DOI] [PubMed] [Google Scholar]
- 6.Natale D.A., Schubert,A.E. and Kowalski,D. (1992) DNA helical stability accounts for mutational defects in a yeast replication origin. Proc. Natl Acad. Sci. USA, 89, 2654–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breslauer K.J., Frank,R., Blocker,H. and Marky,L.A. (1986) Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA, 83, 3746–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin S. and Kowalski,D. (1994) DNA helical instability facilitates initiation at the SV40 replication origin. J. Mol. Biol., 235, 496–507. [DOI] [PubMed] [Google Scholar]
- 9.Miller C.A., Umek,R.M. and Kowalski,D. (1999) The inefficient replication origin from yeast ribosomal DNA is naturally impaired in the ARS consensus sequence and in DNA unwinding. Nucleic Acids Res., 27, 3921–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Natale D.A., Umek,R.M. and Kowalski,D. (1993) Ease of DNA unwinding is a conserved property of yeast replication origins. Nucleic Acids Res., 21, 555–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams D.L. and Kowalski,D. (1993) Easily unwound DNA sequences and hairpin structures in the Epstein–Barr virus origin of plasmid replication. J. Virol., 67, 2707–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carles-Kinch K. and Kreuzer,K.N. (1997) RNA-DNA hybrid formation at a bacteriophage T4 replication origin. J. Mol. Biol., 266, 915–926. [DOI] [PubMed] [Google Scholar]
- 13.Bramhill D. and Kornberg,A. (1988) Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E.coli chromosome. Cell, 52, 743–755. [DOI] [PubMed] [Google Scholar]
- 14.Borowiec J.A. and Hurwitz,J. (1988) Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. EMBO J., 7, 3149–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bode J., Kohwi,Y., Dickinson,L., Joh,T., Klehr,D., Mielke,C. and Kohwi-Shigematsu,T. (1992) Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science, 255, 195–197. [DOI] [PubMed] [Google Scholar]
- 16.Bazar L., Meighen,D., Harris,V., Duncan,R., Levens,D. and Avigan,M. (1995) Targeted melting and binding of a DNA regulatory element by a transactivator of c-myc. J. Biol. Chem., 270, 8241–8248. [DOI] [PubMed] [Google Scholar]
- 17.Charron G., Julien,J.P. and Bibor-Hardy,V. (1995) Neuron specificity of the neurofilament light promoter in transgenic mice requires the presence of DNA unwinding elements. J. Biol. Chem., 270, 25739–25745. [DOI] [PubMed] [Google Scholar]
- 18.Kieffer L.J., Greally,J.M., Landres,I., Nag,S., Nakajima,Y., Kohwi-Shigematsu,T. and Kavathas,P.B. (2002) Identification of a candidate regulatory region in the human CD8 gene complex by colocalization of DNase I hypersensitive sites and matrix attachment regions which bind SATB1 and GATA-3. J. Immunol., 168, 3915–3922. [DOI] [PubMed] [Google Scholar]
- 19.Lin S. and Kowalski,D. (1997) Functional equivalency and diversity of cis-acting elements among yeast replication origins. Mol. Cell. Biol., 17, 5473–5484. [DOI] [PMC free article] [PubMed] [Google Scholar]