Abstract
The ExPASy (the Expert Protein Analysis System) World Wide Web server (http://www.expasy.org), is provided as a service to the life science community by a multidisciplinary team at the Swiss Institute of Bioinformatics (SIB). It provides access to a variety of databases and analytical tools dedicated to proteins and proteomics. ExPASy databases include SWISS-PROT and TrEMBL, SWISS-2DPAGE, PROSITE, ENZYME and the SWISS-MODEL repository. Analysis tools are available for specific tasks relevant to proteomics, similarity searches, pattern and profile searches, post-translational modification prediction, topology prediction, primary, secondary and tertiary structure analysis and sequence alignment. These databases and tools are tightly interlinked: a special emphasis is placed on integration of database entries with related resources developed at the SIB and elsewhere, and the proteomics tools have been designed to read the annotations in SWISS-PROT in order to enhance their predictions. ExPASy started to operate in 1993, as the first WWW server in the field of life sciences. In addition to the main site in Switzerland, seven mirror sites in different continents currently serve the user community.
INTRODUCTION
The Swiss Institute of Bioinformatics (SIB, http://www.isb-sib.ch) is an academic not-for-profit foundation whose mission is to promote research, the development of databanks and computer technologies, teaching and service activities in the field of bioinformatics. One of the SIB's windows to the world is the ExPASy server, which focuses on proteins and proteomics, and provides access to a variety of databases and analysis tools. One of the major assets of ExPASy is the high degree of integration and interconnectivity that it establishes between all the available databases and services. Rather than just making each service accessible in an isolated manner, we put at the disposal of the users different expert views of the complex world of biological data and knowledge.
DATABASES
ExPASy (1,2) is the main host for the following databases that are partially or completely developed at the SIB in Geneva:
The SWISS-PROT knowledgebase (3,4) (http://www.expasy.org/sprot/) is a curated protein sequence database, which strives to provide high quality annotations (such as the description of the function of a protein, its domain structure, post-translational modifications and variants), a minimal level of redundancy and a high level of integration with other databases. SWISS-PROT is supplemented by TrEMBL, which contains computer-annotated entries for all sequences not yet integrated in SWISS-PROT. SWISS-PROT and TrEMBL are maintained collaboratively by the SIB and the European Bioinformatics Institute (EBI).
SWISS-2DPAGE (5) (http://www.expasy.org/ch2d/) is a database of proteins identified on two-dimensional polyacrylamide gel electrophoresis (2D PAGE). SWISS-2DPAGE contains data from a variety of human and mouse biological samples as well as from Arabidopsis thaliana, Escherichia coli, Saccharomyces cerevisiae and Dictyostelium discoideum.
PROSITE (6,7) (http://www.expasy.org/prosite/) is a database of protein domains and families. PROSITE contains biologically significant sites, patterns and profiles that help to reliably identify to which known protein family a new sequence belongs.
ENZYME (8) (http://www.expasy.org/enzyme/) is a repository of information relative to the nomenclature of enzymes.
SWISS-MODEL Repository (9) (http://www.expasy.org/swissmod/smrep.html) is a database of automatically generated structural protein models.
Cross-references
All the databases available on ExPASy are extensively cross-referenced to other molecular biology databases or resources all over the world. SWISS-PROT for example is explicitly cross-referenced (10) to ∼50 different databases specializing in protein and nucleic acid sequences, 3D-structure, organism-specific and genomic information, domain and family signatures, post-translational modifications or proteomics data. Examples for databases currently linked to SWISS-PROT in that manner are EMBL/GenBank/DDBJ, PDB, FlyBase, MGD, MIM, MypuList, SGD, SubtiList, TubercuList, WormPep, ZFIN, InterPro, Pfam, PRINTS, ProDom, PROSITE, SMART, TIGRFAMs, SWISS-2DPAGE, HSSP, MEROPS and REBASE. On average, a SWISS-PROT entry contains 7.8 explicit cross-references to other databases (release 40.43 of 12 February 2003). Literature references for the above-mentioned databases are listed in the SWISS-PROT user manual, (http://www.expasy.org/sprot/userman.html#DR_line).
Complementing these explicit cross-references, so-called ‘implicit links’ to ∼25 additional resources are created on-the-fly by the NiceProt view of SWISS-PROT and TrEMBL entries (see below). This concept is targeted at data collections that do not have their own system of unique identifiers, but can be referenced via identifiers such as SWISS-PROT or EMBL accession numbers, gene names, etc. Examples for databases linked to SWISS-PROT via implicit links are those that are based on SWISS-PROT and provide a specific analytical view of each entry (e.g. ProDom—automatically derived domain views or ProtoMap—a hierarchical classification of all SWISS-PROT entries) and those databases that share some identifier with SWISS-PROT (e.g. GeneCards—information on human genes, accessible by the HUGO approved gene name). Implicit links are a specific feature of ExPASy and are not available on other web servers, or in the SWISS-PROT/TrEMBL data files that can be downloaded by ftp. They greatly enhance database interoperability and strengthen the role of SWISS-PROT as a central hub for the interconnection of biomolecular resources.
Update frequency and download options
SWISS-PROT, PROSITE, ENZYME and SWISS-2DPAGE are updated at a frequency of ∼1–2 weeks.
For all the ExPASy databases, data and associated documentation files can be copied locally by anonymous FTP (ftp.expasy.org). In particular, the different download options for the SWISS-PROT and TrEMBL databases, including the different available subsections, release frequencies and data formats, are documented at http://www.expasy.org/sprot/download.html. Among others, we distribute the files to assemble a non-redundant and complete protein sequence database (ftp://ftp.expasy.org/databases/sp_tr_nrdb/) consisting of three components: SWISS-PROT, TrEMBL and new entries to be later integrated into TrEMBL (known as TrEMBLnew). These files are supplemented by a compilation of sequences for splice variants, reconstructed from the annotations in SWISS-PROT and TrEMBL feature tables. All these files are completely rebuilt every time SWISS-PROT is updated.
A large variety of documents (user manual, release notes, indices, nomenclature documents, etc.) are available with SWISS-PROT; these documents can all be browsed from ExPASy (http://www.expasy.org/sprot/sp-docu.html) and are enhanced by a variety of hyperlinks.
No fees for academic users
The use of all ExPASy databases is free for academic users. However, we implemented in September 1998 a system of annual subscription fee for commercial users of the SWISS-PROT, PROSITE and SWISS-2DPAGE databases. The funds raised are used to bring these databases up-to-date, to keep them up-to-date and to further enhance their quality. Further information on this funding scheme is available at http://www.expasy.org/announce/.
SOFTWARE TOOLS
We have developed, over the years, an extensive collection of software tools, most of which are either targeted toward the access and display of the databases mentioned above, or can be used to analyze protein sequences and proteomics data originating from 2D-PAGE and mass spectrometry experiments. These latter tools can all be accessed from ExPASy (http://www.expasy.org/tools/).
Database query, display and navigation
A variety of query options are available from the home pages of each of the ExPASy databases. These options allow the users to display and retrieve specified subsets of the database. For example, from the home page of SWISS-PROT and TrEMBL, different query forms allow searching by description, accession number, author, citation or by full text search. To complement these options, we have also implemented an SRS (11) server that allows complex searches on any fields of the combination of SWISS-PROT and TrEMBL databases. PROSITE, ENZYME and SWISS-2DPAGE can also be queried using SRS.
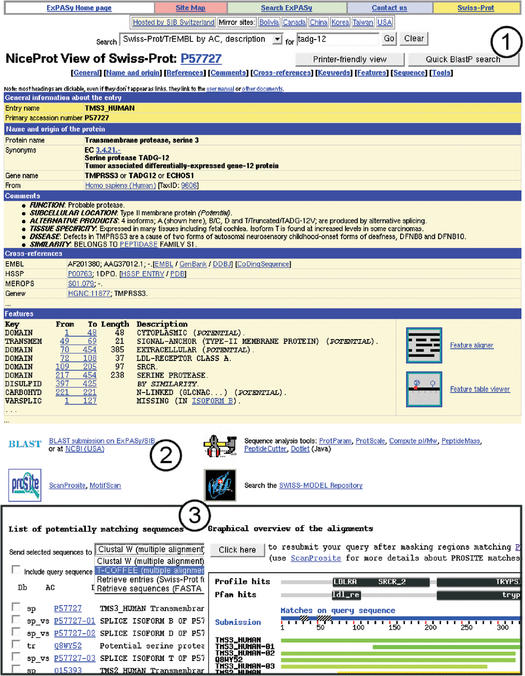
The original flat file format of all ExPASy databases is based on different line types, where a two-letter line code defines the information contained on the rest of that line (e.g. for SWISS-PROT: see the user manual, http://www.expasy.org/sprot/userman.html). This format is easy to parse by computer programs, but not necessarily easy to read for human users. In order to provide a more verbose and user-friendly view of the database entries, we provide for each database, on ExPASy, a ‘nice’ hypertext view, e.g. NiceProt for SWISS-PROT and TrEMBL entries. An example for an entry in the NiceProt view can be seen at http://www.expasy.org/cgi-bin/niceprot.pl?P57727, or in Figure 1. The figure shows parts of that entry in order to illustrate the easy navigation between information contained in the entry itself, the corresponding documentation, remote databases, and the submission forms or results of sequence alignment or other ExPASy analysis tools. Similar views are available for PROSITE (NiceSite and NiceDoc), ENZYME (NiceZyme) and SWISS-2DPAGE (Nice2Dpage).
Figure 1.
The NiceProt view of a SWISS-PROT entry presents its contents in a user-friendly view. Links are provided to >70 databases, a user manual and other documents. NiceProt is also integrated with tools provided on ExPASy and other servers. Excerpts of the view of a sample entry are presented in this figure. A BLASTP similarity search against SWISS-PROT/TrEMBL/TrEMBLnew can be performed with a single click (button 1) on a very fast server (median request time: 6 s). BLAST parameters can also be adjusted by accessing the BLAST page (link 2), which provides a choice of BLASTP or TBLASTN over a choice of databases and subsections. The result page (inset 3) combines a BLASTP search with a motif search in the PROSITE profiles and Pfam HMM domain databases and displays a graphical overview of the matching regions both on the query and on the hit. From there, it is possible to browse matching entries, including splice variants; to rerun a BLAST search after masking regions that match PROSITE or Pfam domains in order to find weaker similarities in other regions and to perform multiple alignments of selected hit sequences.
Swiss-Shop (http://www.expasy.org/swiss-shop/) is an automated sequence alerting system which allows users to obtain new SWISS-PROT entries relevant to their field(s) of interest. Keyword-based and sequence/pattern-based requests are possible. Every time a weekly SWISS-PROT release is performed, all new database entries matching the user-specified search keywords or patterns or the entries showing sequence similarities to the user-specified sequence are automatically sent to the user by email.
Sequence analysis tools
BLAST (12) provides very fast similarity searches of a protein sequence against a protein or nucleotide database. The ExPASy BLAST service is maintained in collaboration with the Swiss EMBnet node on dedicated hardware. The native output of BLAST is extended with several original features (Fig. 1).
ScanProsite (13) scans a sequence against all the patterns, profiles and rules in PROSITE or scans a pattern, profile or rule against all sequences in SWISS-PROT, TrEMBL and/or PDB.
SWISS-MODEL (14,15) is an automated knowledge-based protein modelling server. It is able to build models for the 3D structure of proteins whose sequence is closely related to that of proteins with known 3D structure.
ProtParam calculates physico-chemical parameters of a protein sequence such as the amino acid composition, the pl, the atomic composition, the extinction coefficient, etc.
ProtScale computes and represents the profile produced by any amino acid scale on a selected protein. Some 50 predefined scales are available, such as the Doolittle and Kyte hydrophobicity scale.
RandSeq generates a random protein sequence, based on a user-specified amino acid composition and sequence length.
Sulfinator (16) predicts tyrosine sulfation sites within protein sequences.
Translate translates a nucleotide sequence into a protein in six reading frames.
Proteomics tools
AACompIdent (17) identifies a protein by its amino acid composition.
AACompSim (17) finds for a given SWISS-PROT entry, the database entries which have the most similar amino acid composition.
Compute pI/MW (18) computes the theoretical isoelectric point (pI) and molecular weight (MW) from a SWISS-PROT or TrEMBL entry or for a user sequence.
FindMod (19) predicts potential protein post-translational modifications and potential single amino acid substitutions in peptides. Experimentally measured peptide masses are compared with the theoretical peptides calculated from a specified SWISS-PROT entry or from a user-entered sequence. Mass differences are used to better characterize the protein of interest.
FindPept (20) identifies peptides resulting from unspecific cleavage of proteins by their experimental masses, taking into account artefactual chemical modifications, post-translational modifications and protease autolytic cleavage.
GlycanMass calculates the mass of an oligosaccharide structure.
GlycoMod (21) predicts possible oligosaccharide structures that occur on proteins from their experimentally determined masses. This is done by comparing the mass of a potential glycan to a list of pre-computed masses of glycan compositions.
PeptideCutter predicts potential protease cleavage sites and sites cleaved by chemicals in a given protein sequence.
PeptideMass (22) calculates the theoretical masses of peptides generated by the chemical or enzymatic cleavage of proteins so as to assist in the interpretation of peptide mass fingerprinting.
PeptIdent, TagIdent, MultiIdent (23–25), these three related programs identify proteins using a variety of experimental information such as the pI, the MW, the amino acid composition, partial sequence tags and peptide mass fingerprinting data.
A very important feature of the ExPASy proteomics tools (such as PeptIdent, TagIdent, MultiIdent, PeptideMass, FindPept or FindMod) is that, when performing their computations and predictions, they use the annotations relevant to post-translational modifications and processing, as well as splice variants documented in the SWISS-PROT feature tables.
These tools are all listed on a page on ExPASy (http://www.expasy.org/tools/) that also offers links to many other useful programs for the analysis of protein sequences available elsewhere on the web. We notably have links to the tools provided by our colleagues from the bioinformatics group at ISREC (http://www.isrec.isb-sib.ch) and the Swiss EMBnet node (http://www.ch.embnet.org) in Lausanne. They have developed a BLAST similarity search server, TMpred (to predict transmembrane regions) and interfaces to the SAPS (Statistical Analysis of Protein Sequences), COILS (prediction of coiled coil regions), Clustal and T-Coffee (multiple sequence alignment) programs.
ExPASy AS A PORTAL TO OTHER LIFE SCIENCE RESOURCES
The mass of information available to life scientists on the web has completely changed the way in which biological data is accessed and processed. It has created many opportunities, but also brought new dangers. One of the most critical problems is the difficulty for researchers to distinguish useful and up-to-date sources of information from sites that provide either ‘fossilized’ or low-quality data. To partially address this problem, we have developed a series of lists and tools:
Amos' WWW links page (http://www.expasy.org/alinks.html) is a list that contains links to >1000 information resources for the life sciences. This list is updated very frequently and is organized in a number of sections that correspond to specific topics.
WORLD-2DPAGE (http://www.expasy.org/ch2d/2d-index.html) is a list of all known 2D PAGE database WWW servers and related services.
BioHunt (http://www.expasy.org/BioHunt/) is a service to help search the internet for molecular biology information. BioHunt is built by Marvin, a software robot which automatically roams the web to search and index life science and bioinformatics information. Currently BioHunt indexes ∼35 000 documents.
2DHunt (http://www.expasy.org/ch2d/2DHunt/) is a specialized index for 2D PAGE-related sites.
ExPASy tools page (http://www.expasy.org/tools/), in addition to hosting the above-mentioned tools provided and maintained by the Swiss Institute of Bioinformatics, the tools page serves as a portal to useful web-accessible tools on bioinformatics servers elsewhere. Tools local to the ExPASy server are marked by the ExPASy logo.
List of conferences and events (http://www.expasy.org/conf.html) is a list of conferences and meetings relevant to proteomics, bioinformatics and other domains in the life sciences.
OTHER INTERESTING ExPASy FEATURES
Biochemical pathways (http://www.expasy.org/tools/pathways/) is an indexed, digitized and clickable version of the Boehringer Mannheim's ‘Biochemical Pathways’ poster and is available on the server. It allows the user to navigate through the graphical representation of metabolic pathways and is linked to the ENZYME database.
DeepView (SWISS-PdbViewer) (15) (http://www.expasy.org/spdbv/) is an application running on the Microsoft Windows, Mac, SGI and Linux platforms, offering a wide range of options to visualize and manipulate protein structures. It can also be used as a WWW helper application for the display of PDB formatted entries. Swiss-PdbViewer can be downloaded from ExPASy and complements the aforementioned SWISS-MODEL homology-modeling tool.
LALNVIEW (26) (http://www.expasy.org/tools/lalnview.html) is an application that runs on the Microsoft Windows, Mac and Unix platforms. LALNVIEW is a graphical viewer for pairwise sequence alignments. It can be used to display the results of a pairwise alignment carried out with the SIM (27) software also installed on ExPASy (http://www.expasy.org/tools/sim-prot.html).
2D PAGE: a wide variety of information concerning 2D PAGE is available from ExPASy. This includes the full description of experimental protocols as well as an overview of the Melanie 3 2D PAGE analysis software package. A 2D gel viewer is also available for download.
Protein Spotlight (http://www.expasy.org/spotlight/) is a periodical review centered on a specific protein or group of proteins.
Recreational. One must not forget that science can also have a lighter side. So we hope that users will take the time to take a small pause from the hectic pace of modern research and visit Swiss-Quiz (http://www.expasy.org/swiss-quiz/). With Swiss-Quiz one can have a chance to win some Swiss chocolate (real, not virtual!) after having successfully answered a quiz from the field of molecular biology.
ExPASyBar is a useful navigation bar to the most important databases and tools on ExPASy. ExPASyBar was developed by Martin Hassman from the Institute of Chemical Technology in Prague, in collaboration with the ExPASy team. It is an add-on to the free Mozilla web browser (http://www.mozilla.org), and can be downloaded from http://expasybar.mozdev.org.
MIRROR SITES
Network congestion and resulting slow response times represent a major problem for users in certain parts of the world. To help address this issue, we decided to implement mirror sites of ExPASy in various countries. Such sites can help users to access the ExPASy databases and tools more rapidly in locations that do not have a fast connection to Switzerland. The mirror sites are computers that host exact copies of the information available from the Geneva ExPASy server. They are updated at the same frequency as the main ExPASy site in Switzerland. ExPASy mirror sites are located in academic institutions that have shown an active interest in hosting such sites. As of today, seven sites are operational. The ExPASy mirror sites are located in:
Australia: http://au.expasy.org/ at the Australian Proteome Analysis Facility (APAF), Sydney.
Bolivia: http://bo.expasy.org/ at the Universidad Católica Boliviana (UCB), Cochabamba.
Canada: http://ca.expasy.org/ at the Canadian Bioinformatics Resource (CBR), Halifax.
China: http://cn.expasy.org/ at the Center of Bioinformatics, Peking University, Beijing.
South Korea: http://kr.expasy.org/ at the Yonsei Proteome Research Center.
Taiwan: http://tw.expasy.org/ at the National Health Research Institutes (NHRI), Taipei.
United States: http://us.expasy.org/ at the North Carolina Supercomputing Center (NCSC).
CONCLUSIONS
The team developing ExPASy is committed to bringing to its users top quality information services in the field of proteomics. We hope that in the next years we will be able to add many new features to those that are already available.
We strongly encourage users and providers of related web sites to link to ExPASy. Detailed documentation of how to create html links to the different services of ExPASy is available at http://www.expasy.org/expasy_urls.html.
To keep track of new developments on ExPASy, do not forget to subscribe to Swiss-Flash (http://www.expasy.org/swiss-flash/), a service that allows users to automatically obtain email bulletins that report new and updated ExPASy features.
Acknowledgments
ACKNOWLEDGEMENTS
Finally, we want to thank all the users of ExPASy who, over the years, have sent us feedback that has led to the improvement of existing services and to the development of new ones.
REFERENCES
- 1.Appel R.D., Bairoch,A. and Hochstrasser,D.F. (1994) A new generation of information retrieval tools for biologists: the example of the ExPASy WWW server. Trends Biochem. Sci., 19, 258–260. [DOI] [PubMed] [Google Scholar]
- 2.Bairoch A., Appel,R.D. and Peitsch,M.C. (1997) The ExPASy WWW Server—a tool for proteome research. Protein Data Bank Quart. Newsletter, 81, 5–7. [Google Scholar]
- 3.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M.-C., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O'Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 354–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Donovan C., Martin,M.-J., Gattiker,A., Gasteiger,E., Bairoch,A. and Apweiler,R. (2002) High-quality protein knowledge resource: SWISS-PROT and TrEMBL. Briefings Bioinform., 3, 275–284. [DOI] [PubMed] [Google Scholar]
- 5.Hoogland C., Sanchez,J.-C., Tonella,L., Binz,P.-A., Bairoch,A., Hochstrasser,D.F. and Appel,R.D. (2000) The 1999 SWISS-2DPAGE database update. Nucleic Acids Res., 28, 286–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falquet L., Pagni,M., Bucher,P., Hulo,N., Sigrist,C.J., Hofmann,K. and Bairoch,A. (2002) The PROSITE database, its status in 2002. Nucleic Acids Res., 30, 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sigrist C.J.A., Cerutti,L., Hulo,N., Gattiker,A., Falquet,L., Pagni,M., Bairoch,A. and Bucher,P. (2002) PROSITE: a documented database using patterns and profiles as motif descriptors. Briefings Bioinform., 3, 265–274. [DOI] [PubMed] [Google Scholar]
- 8.Bairoch A. (2000) The ENZYME database in 2000. Nucleic Acids Res., 28, 304–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peitsch M.C. (1997) Large scale protein modelling and model repository. Proc. Int. Conf. Intell. Syst. Mol. Biol., 5, 234–236. [PubMed] [Google Scholar]
- 10.Gasteiger E., Jung,E. and Bairoch,A. (2001) SWISS-PROT: Connecting biological knowledge via a protein database. Curr. Issues Mol. Biol., 3, 47–55. [PubMed] [Google Scholar]
- 11.Etzold T., Ulyanov,A.V. and Argos,P. (1996) SRS: information retrieval system for molecular biology data banks. Methods Enzymol., 266, 114–128. [DOI] [PubMed] [Google Scholar]
- 12.Altschul S.F., Madden,T.L., Schaeffer,A.A., Zhang,J., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gattiker A., Gasteiger,E. and Bairoch,A. (2002) ScanProsite: a reference implementation of a PROSITE scanning tool. Applied Bioinform., 1, 107–108. [PubMed] [Google Scholar]
- 14.Peitsch M.C. (1995) Protein modelling by E-Mail. Biotechnology, 13, 658–660. [Google Scholar]
- 15.Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- 16.Monigatti F., Gasteiger,E., Bairoch,A. and Jung,E. (2002) The Sulfinator: predicting tyrosine sulfation sites in protein sequences. Bioinformatics, 18, 769–770. [DOI] [PubMed] [Google Scholar]
- 17.Wilkins M.R., Pasquali,C., Appel,R.D., Ou,K., Golaz,O., Sanchez,J.C., Yan,J.X., Gooley,A.A., Hughes,G., Humphery-Smith,I. et al. (1996) From proteins to proteomes: large scale protein identification by two-dimensional electrophoresis and amino acid analysis. Bio/Technology, 14, 61–65. [DOI] [PubMed] [Google Scholar]
- 18.Bjellqvist B., Hughes,G.J., Pasquali,C., Paquet,N., Ravier,F., Sanchez,J.-C., Frutiger,S. and Hochstrasser,D.F. (1993) The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis, 14, 1023–1031. [DOI] [PubMed] [Google Scholar]
- 19.Wilkins M.R., Gasteiger,E., Gooley,A.A., Herbert,B.R., Molloy,M.P., Binz,P.-A., Ou,K., Sanchez,J.-C., Bairoch,A., Williams,K.L. and Hochstrasser,D.F. (1999) High-throughput mass spectrometric discovery of protein post-translational modifications. J. Mol. Biol., 289, 645–657. [DOI] [PubMed] [Google Scholar]
- 20.Gattiker A., Bienvenut,W.V., Bairoch,A. and Gasteiger,E. (2002) FindPept, a tool to identify unmatched masses in peptide mass fingerprinting protein identification. Proteomics, 2, 1435–1444. [DOI] [PubMed] [Google Scholar]
- 21.Cooper C.A., Gasteiger,E. and Packer,N. (2001) GlycoMod—a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics, 1, 340–349. [DOI] [PubMed] [Google Scholar]
- 22.Wilkins M.R., Lindskog,I., Gasteiger,E., Bairoch,A., Sanchez,J.-C., Hochstrasser,D.F. and Appel,R.D. (1997) Detailed peptide characterization using PEPTIDEMASS—a World-Wide-Web-accessible tool. Electrophoresis, 18, 403–408. [DOI] [PubMed] [Google Scholar]
- 23.Wilkins M.R., Gasteiger,E., Sanchez,J.-C., Appel,R.D. and Hochstrasser,D.F. (1996) Protein identification with sequence tags. Curr. Biol., 6, 1543–1544. [DOI] [PubMed] [Google Scholar]
- 24.Wilkins M.R., Gasteiger,E., Tonella,L., Ou,K., Tyler,M., Sanchez,J.-C., Gooley,A.A., Walsh,B.J., Bairoch,A., Appel,R.D. et al. (1998) Protein identification with N and C-terminal sequence tags in proteome projects. J. Mol. Biol., 278, 599–608. [DOI] [PubMed] [Google Scholar]
- 25.Wilkins M.R., Gasteiger,E., Wheeler,C., Lindskog,I., Sanchez,J.-C., Bairoch,A., Appel,R.D., Dunn,M.D. and Hochstrasser,D.F. (1998) Multiple parameter cross-species protein identification using MultiIdent—a world wide web accessible tool. Electrophoresis, 19, 3199–3206. [DOI] [PubMed] [Google Scholar]
- 26.Duret L., Gasteiger,E. and Perrière,G. (1996) LALNVIEW: a graphical viewer for pairwise sequence alignments. Comput. Appl. Biosci., 12, 507–510. [DOI] [PubMed] [Google Scholar]
- 27.Huang X. and Miller,W. (1991) A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math., 12, 337–357. [Google Scholar]