Abstract
VADAR (Volume Area Dihedral Angle Reporter) is a comprehensive web server for quantitative protein structure evaluation. It accepts Protein Data Bank (PDB) formatted files or PDB accession numbers as input and calculates, identifies, graphs, reports and/or evaluates a large number (>30) of key structural parameters both for individual residues and for the entire protein. These include excluded volume, accessible surface area, backbone and side chain dihedral angles, secondary structure, hydrogen bonding partners, hydrogen bond energies, steric quality, solvation free energy as well as local and overall fold quality. These derived parameters can be used to rapidly identify both general and residue-specific problems within newly determined protein structures. The VADAR web server is freely accessible at http://redpoll.pharmacy.ualberta.ca/vadar.
INTRODUCTION
Structurally speaking, proteins are perhaps the most complex chemical entities in nature. The large number of atoms, variable composition, convoluted topology and complex surface features make simple descriptions of protein structures almost impossible. The ‘indescribable nature’ of proteins also makes it very difficult to quantitatively assess the quality or correctness of an experimentally determined protein structure. Given the growing number of structures in the Protein Data Bank (PDB) (1) and the growing importance of protein structure in understanding their mechanism, function and evolution, the need to quantitatively describe and evaluate protein structure quality is becoming increasingly important. In response to this need, a number of excellent computer programs have been written specifically to assist structural biologists in this process. These include DSSP (2), WHATIF (3) and PROCHECK (4). Each of these programs specializes in certain areas of protein structure description or assessment. DSSP specializes in automated structure description and has become the gold standard for determining secondary structure, hydrogen bonding and approximate accessible surface area. WHATIF is particularly useful for checking or validating protein geometry and nomenclature with more recent additions allowing more comprehensive structure evaluation. PROCHECK specializes in stereochemical quality evaluation with a particular focus on reporting torsion angle parameters. Each of these programs has their strengths and each can play a vital role in protein structure evaluation or description. However, so far as we are aware, no stand-alone program and certainly no web server calculates or presents all (or nearly all) the DSSP, WHATIF and PROCHECK structural descriptors in a single pass. Furthermore, there are a number of very useful protein structure descriptors and structure checks that have emerged over the past decade that are not reported in DSSP, WHATIF or PROCHECK.
Here we wish to describe a web server, called VADAR, that is able to accept PDB formatted coordinate files and calculate, graph, report and/or evaluate a large number (>30) of key structural parameters for the entire protein as well as numerous structure descriptors for individual atoms, side chains, backbone and residues. VADAR is specifically designed for quantitatively and qualitatively assessing protein structures determined by X-ray crystallography, NMR spectroscopy, 3D-threading or homology modeling.
PROGRAM DESCRIPTION
VADAR (Volume Area Dihedral Angle Reporter) is composed of two parts, a front-end web interface (written in Perl and HTML) and a back-end for calculation (written in C and Fortran). The VADAR server accepts either a PDB formatted file (for newly determined structures) or a PDB accession number (for previously determined structures) as input. The user has a wide choice of radio-button or check-box options for selecting certain parameter sets or activating and de-activating certain calculations or output. Detailed descriptions and references for all of the parameters, criteria and algorithms used in VADAR are provided through an extensive on-line help page. The back-end for VADAR consists of 15 different programs that were newly written, optimized, translated or re-written from previously published algorithms, programs, equations or tables. These include general programs for reading PDB files, as well as specific programs for calculating side-chain and back-bone torsion angles, hydrogen bond energies (2,5), accessible surface area (6), excluded volume (7), secondary structure (2,8,9), beta-turn identity (10), solvation free energy (11,12), secondary structure propensity (13), stereochemical quality (4), 3D profiles (14) as well as numerous routines for global and local statistical analyses.
PROGRAM OUTPUT AND DISCUSSION
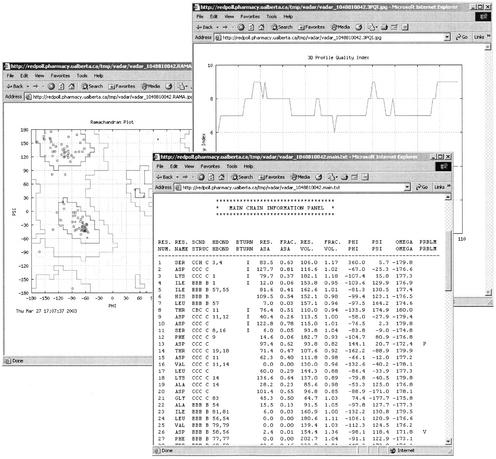
For each input protein structure VADAR automatically generates four sets of detailed, easily printed tables (text format) as well as five sets of scatter plots or line graphs (JPG or PNG format). Each of these tables or graphs is downloadable via a titled hyperlink listed under the VADAR ‘results’ page. A typical VADAR run takes about 5–10 s. Figure 1 provides a sample of the rich graphical and textual output from a standard VADAR run. The first set of tables (MAIN) produced by VADAR uses backbone or main chain coordinates to generate residue-specific data on, secondary structure, turn types, accessible surface area (Å2) fractional ASA, excluded volume (Å3), fractional excluded volume, phi, psi and omega angles. Secondary structure (H=helix, C=coil, B=beta strand) is identified using three different approaches including backbone dihedral angles (8), Ca coordinate masks (9) and hydrogen bonding patterns (2). These three calculations are combined (via a majority vote of the three assignments) to produce a consensus secondary structure assignment. On a test set of 21 high resolution protein structures (with both X-ray and NMR data) these assignments were found to agree well (>90% concordance) with the original authors' assignments, with NMR secondary structure assignments and with DSSP secondary structure designations. Beta-turn classification and identification is done according to the method of Wilmot and Thornton (10) with the added requirement that beta-turns cannot be placed wholly within previously identified helices or beta strands.
Figure 1.
A screenshot montage of VADAR output for thioredoxin (2TRX) showing an example of the Ramachandran plot, the MAIN (main chain) tables and the 3D profile plot (quality index). 2TRX is a good example of a high quality, high resolution structure.
Accessible surface areas (both fractional and absolute) are calculated using the ANAREA program using a 1.4 Å probe radius (6). ASA is highly dependent on the choice of atomic or Van der Waals radii. Different authors and sources use different radii and VADAR provides four choices. Shrake and Rupley's (15) atomic parameters are used as default values. The fractional residue ASA (for a user-chosen set of radii) is determined by dividing the observed ASA (Å2) for a given residue by the calculated ASA for that residue in an extended Gly-Xaa-Gly tripeptide. VADAR uses pre-calculated tables of extended-residue areas derived from each of the four program options to ensure the fractional areas are consistent with the user-chosen option.
VADAR reports ASA values both for the whole residue and for side chains. ASA values are also calculated for polar (N, O, S) atoms, charged atoms (N+, O−) and for non-polar atoms (C) to permit the calculation of polar, charged and non-polar surface area. These ASA values can be quite useful in structure assessment and in thermodynamic calculations.
Excluded volume is calculated using the Vornoi polyhedra method of Richards (7). Excluded volume represents the volume occupied by a residue as defined by its atomic radii and its nearest neighbors. Normally, if the protein is efficiently packed, all residues should have fractional volumes close to 1.0±0.1 (Table 1). A residue located in an interior cavity (or which has been improperly placed) will typically have a fractional excluded volume >1.20. A residue located in a compressed region or a poorly refined region of a protein structure will typically have a fractional volume <0.80. Excluded volume is a good way of finding cavities, water-binding pockets, excessive atomic overlaps or other problem areas in a protein structure. In VADAR, cavities and compressions are called ‘packing defects’.
Table 1. Limits and variation for structural assessment parameters.
| High resolution structures | Misfolded structures | |
|---|---|---|
| Fractional ASA >1.0 | 0.28% | 0.30% |
| Fractional volume | 0.98±0.11 | 1.00±0.16 |
| Fractional volume >1.2 | 3.95% | 9.67% |
| Fractional volume <0.8 | 1.15% | 4.09% |
| Packing defects | 5.42% | 14.26% |
| Stereo index | 8.76±0.64 | 7.58±1.48 |
| Stereo index <7 | 2.6% | 24.33% |
| 3D profile score | 6.32±1.57 | 5.02±2.16 |
| 3D profile score <5 | 15.34% | 46.96% |
| Buried charges | 1.46% | 3.49% |
| Number of residues | 2530 | 1003 |
In addition to providing a wide range of residue specific structure descriptors, data from the MAIN tables can also be used to check for bifurcated hydrogen bonds, the existence of rare beta-turns, distorted backbone angles (omega angles <170°, positive phi angles), the presence of cis-peptide bonds, evidence of buried charges (ASA of charged amino acids near 0) or unusual cavities (residue fractional volume >1.20) or residue compressions (residue fractional volume <0.80). Outliers or possible problem residues are flagged in the rightmost column of the MAIN table with appropriately referenced single letter designations (P for phi/psi outliers, O for omega outliers, C for cis peptide bonds, V for volume outliers and A for ASA outliers). Outliers are identified using published limits (4) or data derived from our own analyses (Table 1, vide infra).
The second set of tables (SIDE) produced by VADAR reports similar residue-specific data for side chain atoms, including side chain hydrogen bonds and side chain chi-1 angles. This data allows users to evaluate and identify side chain anomalies that may not be obvious from main chain data. The third set of tables (HBOND) reports data (energy, bond length, residue label, angle, donor, acceptor) on all identified pairs of hydrogen bonds (backbone and side chain). Hydrogen bonds are identified and their energies calculated using the method of Kabsch and Sander (2) with modifications suggested by Baker and Hubbard (5).
The fourth and final set of tables (STATS) compiles the residue- or atom-specific data from the SIDE, HBOND and MAIN to generate global statistics that can be used to evaluate the structure's overall quality. Averages, standard deviations and values relative to known high-resolution (or idealized) structures are calculated and presented for hydrogen bond lengths, bond angles, helix dihedral angles, polar, charged and non-polar accessible surface area, excluded volume, and other parameters. Many of the values, limits and standard deviations quoted in the STATS tables were derived from well-known literature sources (4,7,16) and are individually referenced in each STATS table. However, some of the values pertaining to volume, ASA, charge burial, stereo-quality indices and 3D profile indices are unique to VADAR. To derive the limits and variances for these parameters we analyzed a set of 21 high resolution (<1.8 Å) structures as well as seven misfolded, poorly resolved or mis-traced structures (obsolete PDB entries). The PDB accession numbers and/or file hyperlinks for all 28 proteins are available at the VADAR help page. The results of these analyses are presented in Table 1 and clearly show the significant differences (2–10-fold) in many of these calculated parameters between ‘good’ and ‘bad’ structures. These data also provide a good rationale for the limits chosen to identify possible outliers in a standard VADAR analysis.
As indicated earlier, the STATS tables also display other calculated indices regarding the quality of the structure or viability of the fold. These quality indices attempt to summarize the quality of the input protein structure in two ways. One is a stereochemical/packing quality assessment and the other is a threading or 3D profile assessment. The stereochemical/packing quality index categorizes phi/psi and omega trends according the criteria given by Morris et al. (4). It also includes the presence of packing defects (excessively large cavities or atomic overlaps) as part of the quality score. These stereochemical quality indices allow specific ‘problem’ residues to be rapidly identified (i.e. residues with scores <7, which are also marked with an asterisk in the STATS table). High quality or high resolution structures typically have scores close to 9 for all residues (Table 1). The second quality index uses threading or a variant of the 3D-profile method of Luthy et al. (14) to assess the local environment, packing and hydrophobic energy for the given structure. The threading score also includes the secondary structure propensity (calculated via the GOR method) as compared to the observed secondary structure. Typically these threading or 3D-profile quality indices range between 5 and 8 (Table 1). Values that are significantly lower (<5, which are also marked with an asterisk in the STATS table) indicate possible problems with the local structure or local fold.
In addition to these tabular data sets, VADAR also uses GNU-PLOT to generate a series of scatter plots and line graphs from selected VADAR output. These include graphs corresponding to fractional ASA, fractional volume, the two quality indices and a Ramachandran distribution plot. These five graphs, which highlight outliers as well as upper/lower limits for specific values, are provided as aids for more rapid visual assessment of protein structure quality. Users have the option of saving these graphs as either fixed width or variable width (constant pixels/residue) images in JPG or PNG format.
In summary, VADAR is a comprehensive web server for protein structure evaluation that both complements and adds to existing structure assessment programs. VADAR represents a compilation of >30 key structural parameters derived from 15 well-known algorithms or previously published techniques for quantitatively evaluating protein structures. A large number of these algorithms have been re-written and optimized to improve their results and facilitate rapid on-line calculation. VADAR should be particularly useful for evaluating newly determined X-ray, NMR or homology modeled protein structures. The VADAR web server is freely accessible at http://redpoll.pharmacy.ualberta.ca/vadar.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank F.M. Richards (Yale University) for his advice and assistance in making the ANAREA and VOLUME source code available. Funding for this project was provided by the Protein Engineering Network of Centres of Excellence (PENCE Inc.) and the Alberta Heritage Foundation for Medical Research.
REFERENCES
- 1.Westbrook J., Feng,Z., Chen,L., Yang,H. and Berman,H.M. (2003) The Protein Data Bank and structural genomics. Nucleic Acids Res., 31, 489–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- 3.Vriend G. (1990) WHAT IF: A molecular modeling and drug design program. J. Mol. Graph., 8, 52–55. [DOI] [PubMed] [Google Scholar]
- 4.Morris A.L., MacArthur,M.W., Hutchinson,E.G. and Thornton,J.M. (1992) Stereochemical quality of protein structure coordinates. Proteins, 12, 345–364. [DOI] [PubMed] [Google Scholar]
- 5.Baker E.N. and Hubbard,R.E. (1984) Hydrogen bonding in globular proteins. Prog. Biophys. Mol. Biol., 44, 97–179. [DOI] [PubMed] [Google Scholar]
- 6.Lee B. and Richards,F.M. (1971) The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol., 55, 379–400. [DOI] [PubMed] [Google Scholar]
- 7.Richards F.M. (1977) Areas, volumes, packing and protein structure. Annu. Rev. Biophys. Bioeng., 6, 151–176. [DOI] [PubMed] [Google Scholar]
- 8.Levitt M. and Greer,J. (1977) Automatic identification of secondary structure in globular proteins. J. Mol. Biol., 114, 181–239. [DOI] [PubMed] [Google Scholar]
- 9.Richards F.M. and Kundrot,C.E. (1988) Identification of structural motifs from protein coordinate data: secondary structure and first-level supersecondary structure. Proteins, 3, 71–84. [DOI] [PubMed] [Google Scholar]
- 10.Wilmot C.M. and Thornton,J.M. (1988) Analysis and prediction of the different types of beta-turn in proteins. J. Mol. Biol., 203, 221–232. [DOI] [PubMed] [Google Scholar]
- 11.Chiche L., Gregoret,L.M., Cohen,F.E. and Kollman,P.A. (1990) Protein model structure evaluation using the solvation free energy of folding. Proc. Natl Acad. Sci. USA, 87, 3240–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenberg D, and McLachlan,A.D. (1986) Solvation energy in protein folding and binding. Nature, 319, 199–203. [DOI] [PubMed] [Google Scholar]
- 13.Garnier J., Osguthorpe,D.J. and Robson,B. (1980) Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J. Mol. Biol., 120, 97–120. [DOI] [PubMed] [Google Scholar]
- 14.Luthy R., Bowie,J.U. and Eisenberg,D. (1992) Assessment of protein models with three-dimensional profiles. Nature, 356, 83–85. [DOI] [PubMed] [Google Scholar]
- 15.Shrake A. and Rupley,J.A. (1973) Environment and exposure to solvent of protein atoms. Lysozyme and insulin. J. Mol. Biol., 79, 351–371. [DOI] [PubMed] [Google Scholar]
- 16.Miller S., Janin,J., Lesk,A.M. and Chothia,C. (1987) Interior and surface of monomeric proteins. J. Mol. Biol., 196, 641–656. [DOI] [PubMed] [Google Scholar]