Abstract
The web resource Regulatory Sequence Analysis Tools (RSAT) (http://rsat.ulb.ac.be/rsat) offers a collection of software tools dedicated to the prediction of regulatory sites in non-coding DNA sequences. These tools include sequence retrieval, pattern discovery, pattern matching, genome-scale pattern matching, feature-map drawing, random sequence generation and other utilities. Alternative formats are supported for the representation of regulatory motifs (strings or position-specific scoring matrices) and several algorithms are proposed for pattern discovery. RSAT currently holds >100 fully sequenced genomes and these data are regularly updated from GenBank.
INTRODUCTION
Despite the essential role played by non-coding sequences in transcriptional regulation, genome annotations usually focus on identifying the genes and predicting their function through sequence similarity searches. The services offered by most genome centers are restricted to analysis of coding and peptidic sequences. The web resource Regulatory Sequence Analysis Tools (RSAT) is dedicated to the analysis of the other part of the genomes: the non-coding sequences. It proposes a collection of modular tools which can be combined in different ways to predict regulatory elements. Three main scenarios can be handled: (i) starting from a set of co-regulated genes, retrieve their upstream sequences and detect over-represented motifs, which might be responsible for their co-regulation ( pattern discovery); (ii) predict the location of binding sites for a known transcription factor in a given sequence ( pattern matching); (iii) starting from the known consensus pattern for a given transcription factor, scan all upstream sequences of a selected genome in order to predict putative target genes (genome-scale pattern matching).
TASKS AND PROGRAMS
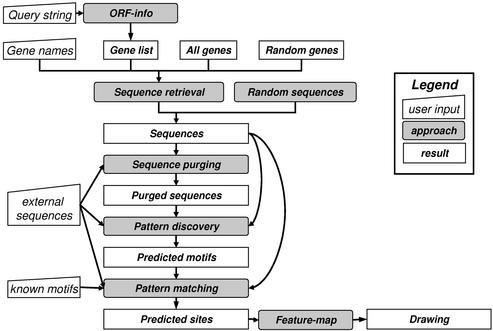
The procedures currently supported by RSAT are summarized in Table 1. These procedures are linked in a pipeline as illustrated in Figure 1. In the following we describe different tasks and the programs that are most appropriate for performing them.
Table 1. Summary description of the tools.
| Task | Program name | Input | Output | Description |
|---|---|---|---|---|
| Sequence retrieval | retrieve-seq | gene names | sequences | Given a set of gene names, returns upstream, downstream or unsplied ORF sequences. The user defines the limits relative to the ORF start. Segments overlapping an upstream ORF can be excluded or included. |
| Pattern discovery | oligo-analysis | sequences | over-represented oligonucleotides | Analyze oligonucleotide occurrences in a set of sequences, and detects over-rpresented oligonucleotides. Various background models and scoring statistics are supported. |
| dyad-analysis | sequences | over-represented dyads | Detects over-represented dyads (spaced pairs of oligonucleotides) within a set of sequences. | |
| consensus | sequences | PSSM (position-specific scoring matrix) | Detects shared motifs in unaligned sequences on the basis of a greedy algorithm. Developed by Jerry Hertz. | |
| gibbs | sequences | PSSM | Detects shared motifs in unaligned sequences on the basis of a Gibbs sampling strategy. Developed by Andrew Neuwald. | |
| Pattern matching | dna-pattern | sequences+multiple patterns (string description) | matching positions in input sequences | String matching program specialized for DNA sequences. IUPAC code for partially specified nucleotides is supported, as well as regular expressions. Several patterns can be searched simultaneously in several sequences, allowing a fast detection of multiple features. Searches can be performed on a single or both strands. |
| patser | sequences+one pattern (PSSM) | matching positions in input sequences | Pattern matching program based on a position-specific scoring matrix description of the patterns. Developed by Jerry Hertz. | |
| genome-scale-dna-pattern | multiple patterns (string description) | matching positions in all upstream sequences | Pattern matching with dna-pattern, applied to all genes (upstream or downstream sequences) of a selected organism | |
| genome-scale-patser | single pattern (PSSM) | matching positions in all upstream sequences | Pattern matching with patser, applied to all genes (upstream or downstream sequences) of a selected organism | |
| Drawing | feature-map | matching positions | drawing | Draws a map with the results of pattern matching programs. Several sequences can be represented in parallel, allowing visual comparison of matching positions. |
| XYgraph | numbers | drawing | Draws a 2-D graph from a table of numerical data. | |
| Utilities | ORF information | gene names | genes | Selects genes whose identifier, name or description matches a list of query strings. Partial matches are supported. |
| pattern-assembly | string patterns | alignment | Aligns a set of strongly overlapping patterns (oligos or dyads). | |
| purge-sequences | sequences | sequences | Discards large repetitive fragments from a sequence set. Program developed by Stefan Kurtz. | |
| convert-seq | sequences | sequences | Interconversions between different sequence formats. | |
| random-seq | sequences | Generates random sequences. Different probabilistic models are proposed (equiprobable nucleotides, specific alphabet utilization, Markov chains). | ||
| random-genes | organism | genes | Selects a random set of genes. |
Figure 1.
Flow chart of the Regulatory Sequence Analysis Tools.
Sequence retrieval
The simplest input for RSAT is a list of gene names. Using this list the retrieve-seq program returns upstream, downstream or unspliced ORF sequences (introns and spliced ORFs will soon be supported). The user can specify the left and right limits of the sequences to be retrieved. Default values have been selected for each genome, depending on the average size of the intergenic regions and mechanisms of regulation. Upstream sequences can be retrieved over a constant size, but an option also allows to clip them in order to avoid the inclusion of coding sequences from upstream ORFs.
Transcription factor binding sites
The specificity of a transcription factor can be described by a pattern. Two alternative formats are currently used to describe regulatory signals: strings (including the IUPAC alphabet for ambiguous nucleotides) or position-specific scoring matrices (PSSM) (1).
Pattern matching
When the regulatory pattern is known (e.g. the consensus binding sequence for a given transcription factor), one may wish to locate its occurrences, in order to identify putative transcription factor binding sites in upstream sequences of a set of genes. Patterns can be collected from the literature or obtained from specialized databases (2–4). String-based pattern matching is performed with the program dna-pattern. This program supports the IUPAC degenerate alphabet, as well as regular expressions, which allow the specification of spaces of variable length. Patterns can be searched on either one or both strands. A matrix-based pattern matching procedure, patser, developed by Jerry Hertz (5,6), has been integrated to the web interface.
Pattern discovery
Given a set of co-regulated genes, pattern discovery programs can be used to detect over-represented motifs in their upstream regions. This is particularly useful for the prediction of regulatory motifs from clusters of co-expressed genes, such as those obtained from microarray data or other high-throughput methods. Several algorithms for pattern discovery are supported. The program oligo-analysis (7) analyzes oligonucleotide occurrences and returns those that are statistically over-represented (Table 2A).
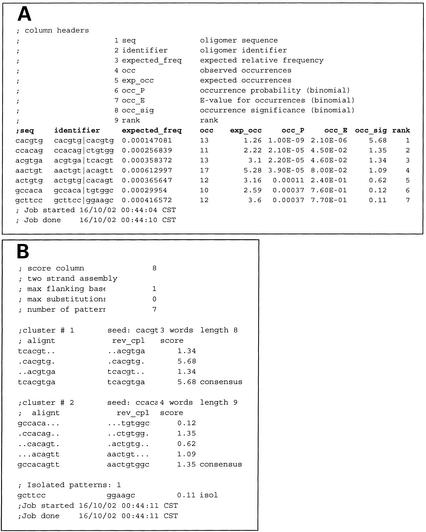
Table 2. Output of oligo-analysis applied to a set of 12 yeast genes regulated by methionine (SAM2, MET6, MUP3, MET30, MET3, MET14, MET1, SAM1, MET17, ZWF1 and MET2).
Upstream sequences were retrieved >800 bp from the start codon. (A) Over-represented oligonucleotides. Each row represents one pattern, i.e. a hexanucleotide with its reverse complement. Each pattern has a specific prior probability (exp_freq) defined on the basis of the selected background model (e.g. intergenic sequences from S. cerevisiae). For each pattern, the number of occurrences (occ) is compared to the random expectation (exp_occ), and the P-value is calculated (occ_P). The E-value (occ_E) is obtained by multiplying the P-value (occ_P) by the number of patterns considered, in order to correct for multi-testing. In this case, no less than 2080 patterns were tested, so the correction is important. The significance index (occ_sig) equals -log (E-value). Over-represented patterns have positive occ_sig values. In the sequences analyzed here, no more than seven patterns were significant among the 2080 possiblities. Notice that some of the patterns strongly overlap with each other. (B) The seven patterns from Table A were aligned with the program pattern-assembly, resulting in two clusters of hexanucleotides, each forming a larger consensus. The most significant result, TCACGTGA, is the consensus of the Met4p/Cbf1p/Met28p complex. The second consensus, AACTGTGGC, corresponds to the binding site for Met31p.
Despite it simplicity, this program has proven to be very efficient for the detection of regulatory motifs in the yeast Saccharomyces cerevisiae. However, some motifs escape detection, because they take the form of a spaced dyad, i.e. a pair of very short oligonucleotides separated by a region of fixed length but variable content. A second program, dyad-analysis (8), specifically detects such spaced motifs, which are typical of many bacterial transcription factors, and of the fungal binuclear zinc cluster proteins. String-based pattern discovery programs generally return several oligonucleotides or dyads, which can be assembled with the program pattern-assembly, to yield larger and/or partially degenerate motifs (Table 2B). Two matrix-based pattern discovery programs, Andrew Neuwald's gibbs sampler (9) and Jerry Hertz's consensus (5,6), are also available.
The strength of string-based pattern discovery methods is their very low rate of false positives and the fact that they are able to return multiple motifs when a set of genes is regulated by several factors. This is illustrated by the example in Table 2B, where the analysis of 10 methionine-responsive genes led to the detection of two distinct patterns, corresponding to the binding sites of Met4p and Met31p, respectively. Matrix-based programs return a more refined description of pattern degeneracy, but have the drawback of always returning an answer, even when random sequences are submitted.
Genome-scale pattern matching
Pattern matching can be applied to the full set of upstream sequences in a genome, in order to predict genes possibly regulated by a given transcription factor. It should be noted that the simple presence of a motif in a given upstream region is generally not sufficient to predict regulation. Indeed, given the short size of the motifs and the large size of the genomes, hundreds, or even thousands of matches could be returned by chance alone. Predictions can be improved by detecting multiple binding sites, either for the same transcription factor, or for combinations of several different transcription factors.
Feature map drawing
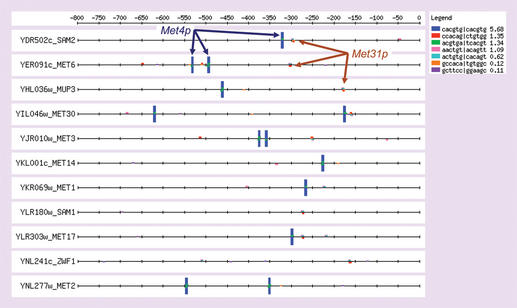
The results obtained by pattern matching can be displayed graphically, in the form of a feature map (Fig. 2). In this map, each motif is represented by a box painted in a different color, whose height is proportional to the statistical significance of the pattern. Feature maps are not only useful for illustrative purposes, they can also reflect additional properties of the discovered motifs such as a conserved position relative to the start codon, a distal or proximal location, the pairing of heterologous motifs and so on.
Figure 2.
Feature map of the patterns discovered in Table 2. Each box corresponds to one hexanucleotide. Transcription factor binding sites are generally revealed as combinations of boxes, which reflect the fact that the consensus is >6 nt. The blue box corresponds to CACGTG, the core of Met4p binding site. Some examples of the smaller boxes, corresponding to putative Met31p binding sites, are indicated. Notice that putative Met31p binding sites are often associated to putative Met4p binding sites, suggesting an interaction between the two factors.
Random sequences and random gene selections
Random sequences are useful for performing negative controls. Indeed, some programs present the inconvenient of systematically returning an answer, even when the submitted sequence set contains no biologically significant features. The program random-seq generates random DNA sequences on the basis of various probabilistic models (independent nucleotides, Markov chains).
Another program, random-genes, selects random sets of genes for a given organism. Random gene selections provide a very stringent test for pattern discovery programs. Indeed, although each selected gene is likely to have some regulatory elements, there is no reason for the selected group, as a whole, to be co-regulated, and a good pattern discovery program should thus generally return a negative answer or motifs with low significance.
WEB INTERFACE
The originality of the RSAT resource is that it provides an integrated approach for tackling a variety of questions about regulatory sequences. The tools are integrated into a pipeline (Fig. 1), but can also be used individually by filling the forms with data from external sources. This includes the uploading of large sequence files.
The web interface has been designed so as to allow ready access to the tools by non-specialists. Default parameters have been defined on the basis of previous experience. In addition, a user manual provides a detailed description of the options for each program. Moreover, a series of tutorials are available for the step-by-step initiation of first-time users.
PERSPECTIVES
The web tools presented here perform predictions of regulatory elements using several approaches under various circumstances. The approaches used have strong points as well as limitations of which one should be well aware. Any predictive method will unavoidably return false positives and/or miss some genuine regulatory patterns. Until now, most methods have been optimized and validated in microbial model organisms (yeast and bacteria). Whether these approaches can be extended to higher organisms is still an open question. Currently we are evaluating the applicability of RSAT for the detection of regulatory elements in the genomes of multicellular organisms. To perform such detection successfully, new methods, based on comparative genomics, might be required (reviewed in 10).
Acknowledgments
ACKNOWLEDGEMENTS
I acknowledge support from the Action de Recherches Concertées de la Communauté Française de Belgique (contract ARC-97/01-211). I am grateful to Jean Richelle for the system administration and to Shoshana Wodak who helped to improve this manuscript. The RSAT project was originated at the Universidad Nacional Autonoma de Mexico, in the laboratory of Julio Collado-Vides, to whom I am thankful for past and present collaboration.
REFERENCES
- 1.van Helden J., Andre,B. and Collado-Vides,J. (2000) A web site for the computational analysis of yeast regulatory sequences. Yeast, 16, 177–187. [DOI] [PubMed] [Google Scholar]
- 2.Zhu J. and Zhang,M.Q. (1999) SCPD: a promoter database of the yeast Saccharomyces cerevisiae. Bioinformatics, 15, 607–611. [DOI] [PubMed] [Google Scholar]
- 3.Matys V. Fricke,E., Geffers,R., Gossling,E., Haubrock,M., Hehl,R., Hornischer,K., Karas,D., Kel,A.E., Kel-Margoulis,O.V. et al. (2003) TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res., 31, 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salgado H., Santos-Zavaleta,A., Gama-Castro,S., Millan-Zarate,D., Diaz-Peredo,E., Sanchez-Solano,F., Perez-Rueda,E., Bonavides-Martinez,C. and Collado-Vides,J. (2001) RegulonDB (version 3.2): transcriptional regulation and operon organization in Escherichia coli K-12. Nucleic Acids Res., 29, 72–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hertz G.Z., Hartzell,G.W.d. and Stormo,G.D. (1990) Identification of consensus patterns in unaligned DNA sequences known to be functionally related. Comput. Appl. Biosci., 6, 81–92. [DOI] [PubMed] [Google Scholar]
- 6.Hertz G.Z. and Stormo,G.D. (1999) Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics, 15, 563–577. [DOI] [PubMed] [Google Scholar]
- 7.van Helden J., Andre,B. and Collado-Vides,J. (1998) Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol., 281, 827–842. [DOI] [PubMed] [Google Scholar]
- 8.van Helden J., Rios,A.F. and Collado-Vides,J. (2000) Discovering regulatory elements in non-coding sequences by analysis of spaced dyads. Nucleic Acids Res., 28, 1808–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neuwald A.F., Liu,J.S. and Lawrence,C.E. (1995) Gibbs motif sampling: detection of bacterial outer membrane protein repeats. Protein Sci., 4, 1618–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Helden J. (2003) Prediction of transcriptional regulation by analysis of the non-coding genome. Curr. Genomics, 4, 217–224. [Google Scholar]