Abstract
We describe an integrated tool-box to identify regulatory RNA elements. The RNA analyzer collects general and specific information on any submitted RNA sequence or batch of sequences in FASTA format. It determines and rapidly scans the different regions of an RNA (including 5′ UTR, CDS, 3′ UTR in mRNA) and screens for specific RNA signals (in each of these regions, e.g. polyA-site, AU rich region etc. in 3′ UTR). It runs a fast folding RNA routine to provide an overview of the RNA fold. Furthermore it analyzes structure content, fold energy and stem loops. In addition, consensus templates are used to determine whether there are any functional structures present for translational control (template: IRE), structured RNA (template: tRNA consensus) or catalytic RNA (template: trans-splicing RNA), giving indications as to how well the structures found match to these templates. The tool box has been implemented as a WWW server at http://wb2x01.biozentrum.uni-wuerzburg.de/.
INTRODUCTION
A number of approaches and modules are available to analyze and identify regulatory RNA motifs and structures (reviewed in 1).
For specific regulatory elements a number of programs are available (e.g. IREs and tRNA scan). However, it is often desirable to get a comprehensive overview on the content of regulatory elements in a given RNA sequence. Different approaches are feasible for this including entropy analysis (2) or Hidden Markov models (3) [also applied in promotor analysis and gene identification (4)]. However, the information is often quite unspecific, for example the entropy based approach mentioned above indicates only a probability for the RNA region in question to contain a non-random structure.
Instead we describe and strive here to get a fast responding program suite. A number of different routines are bundled for this including custom written new routines for the effort. The approach is heuristic and application oriented and rapidly scans for a number of specific regulatory RNA elements and alerts the user to the existence of potential further regulatory structures. To give guides in the latter case, our scanning routines indicate the potential for three categories: potential catalytic element, potential structural motif or potential translational element. Decisions for each of these categories are again based on the presence of small motifs or parts of higher order, more complex structures to alert the user in which direction to examine the structures and at which specific parts of the RNA the motif or potential motif is located.
MATERIALS AND METHODS
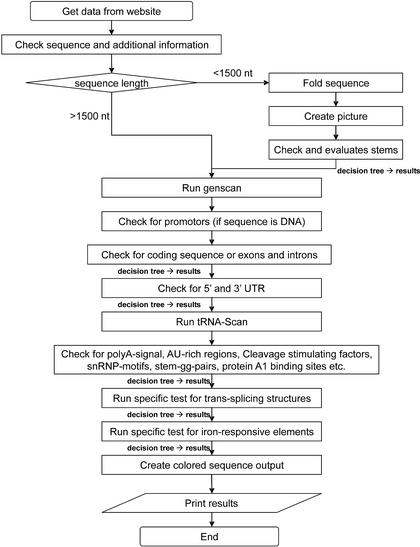
For the presence or absence of each motif the program rapidly goes through different rule-based decision trees as indicated in the program flow (Fig. 1). For specific or more complex regulatory motifs, individual subroutines are called which again use either rule-based motif scans or other approaches which proved to be highly specific for the motif in question.
Figure 1.
The program flow is shown. Note that in the different check boxes additional information, different stems and a list of different regulatory motifs are analyzed by several subroutines (not shown in detail).
General analysis routines
First the sequence format is checked to determine and whether the sequence is DNA or RNA. We included RNA folding routines as used and implemented by Stiegler and Zuker (5,6) and made available through the Vienna RNA package (7). Up to 1500 nucleotides, each RNA is folded completely. The folding created is printed as a picture and is given, in addition, in bracket notation. Moreover, stems are checked and evaluated. Reading frames are next detected using the Genscan (8) sub-routine including identification of promoters, exons and UTRs. Next, tRNA scan detects such tRNA structures (see below) after which a number of simpler motifs are tested. This included motif tests as detailed in Table 1 and results. Threshold for ARE detection was set low enough to identify the ARE in well known ARE sequences such as interferon alpha 21 (GenBank identifier M 15330). Reading frame threshold in the Genscan routine was set to a minimum length of 100 amino acids as this is a typical threshold used in many genome projects. These and any other thresholds can easily be modified if so desired by the user in his copy of the source code (which is available on request from the authors).
Table 1. Output overview and examples.
| Analysis of | Analysis done and output |
|---|---|
| Complete sequence: Alert: ‘some info is only avail up to 1500 nt’ | Alert if sequence is longer than 1500 nucleotides |
| Complete sequence: determination of RNA/DNA | Scan nucleotides. Prediction whether molecule is DNA or RNA. |
| Complete sequence: rRNA pre-screen | If there are more than 10 stems indicate ‘Highly structured RNA, could this be a ribosomal RNA?’ |
| Complete sequence: RNA but no coding sequence | Sequence is scanned in 150 nt steps and the number of stems is given. |
| Complete sequence: Alert for highly structure RNA: (in RNA molecules without exons the whole sequence is folded in 150 nt sections) | If there are three or more stems per 150 nt follows a prompt: ‘Three or more stem loops in this region! This is a highly structured region. Please check wheter tRNA, rRNA or another highly structured RNA is encoded here!’ |
| Complete sequence: Hidden structures | If there is more than one stem per 60 nucleotides (regarding the complete sequence) prompt: ‘The sequence seems to contain a lot of secondary structure. If the RNA structure search below does not find a result it might be interesting to have a closer look at the structures. You might find it useful to use a reference book [e.g. (1)]’ |
| Promotor | Start and End are indicated |
| UTR | Start, End, stems and their energy are indicated |
| 5′ UTR | is indicated |
| Alert for potential catalytic RNA if no exons are present, but molecule is RNA and Sm-sites have been found | ‘As I could not detect a coding sequence on this RNA, but there are 1 or more sn-RNP motifs (sm-sites), it might be possible, that this is a catalytic RNA’ |
| Protein A1 binding site | Start, mismatches, and exact sequence are given |
| Sm-Site or snRNP binding motif | Start and End, quality and whether an snRNP or sm-site is indicated |
| Exons | Start and End and whether coding region is present |
| Protein prediction | Indication of coding sequence/protein sequence. Call to the structural domain server AnDom (19) |
| 3′ UTR | if more than 3, 5 stems are localized here, a message indicates ‘potential stability elements might be located in this 3′ UTR’ |
| Polyadenylation-signal | Start and End are indicated |
| AU-rich region ARE | Start and End are indicated |
| CstF | Cleavage stimulation Factor binding region: Start, mismatches |
| GG-pairs | Position, rev-response element like feature |
| Specific regulatory RNA elements or structures: tRNA | The complete tRNA structure and a number of describing notes on it are given |
| Specific regulatory RNA elements or structures: Trans-splicing donor structure | Complete regulatory element is identified, calculated and indicated |
| Specific regulatory RNA elements or structures: Iron-responsive element (IRE) | Complete regulatory element is identified, calculated and indicated |
Subroutines for identification of specific functional RNAs
IREs were detected by the program by Dandekar et al. (9). It combines motif and secondary structure searches with a scoring scheme. The implemented version now includes subsequent modifications (10) and improved folding energy calculation (programmed by P.B., 2001). All known IREs are successfully identified by this routine and it has successfully identified several new IREs in other mRNAs.
Identification of trans-splicing structures followed the program by Dandekar and Sibbald (11) including subsequent modifications (12) and again improved folding energy calculation (programmed by P.B., 2001).
tRNA identification uses the program tRNA-scan-SE (13). TRNA-scan-SE identifies 99–100% of transfer RNA genes in DNA sequence while giving less than one false positive per 15 Gb. It applies RNA covariance models, using probabilistic secondary structure profiles based on stochastic context-free grammars (13).
Program package
For the program package a web interface was written and the server is available for the community. It runs on an AMD athlon processor machine. In addition, source code and simple installation protocol are available for Linux on request from the authors.
RESULTS AND DISCUSSION
Query
A query is posted by simply pasting the sequence into the query window (accepted formats: Raw, FASTA, file of FASTA sequences). Run time scales quadratically with sequence length [O(n2)] including the plot of the RNA fold (up to 1500 nt) and increases only linearly thereafter.
Data
Any given RNA sequence can be analyzed. Up to 1500 bp the complete folding is calculated.
However, also a complete database file or a chunk of genomic DNA can be searched if provided in FASTA format.
RNA analysis routines
Figure 1 shows the flow of the program. Starting from the input and folding sequences up to 1500 nt, after a general analysis more and more specific motifs are analyzed by different subroutines, all results are collected and a coloured sequence output is created.
Analysis results
The various RNA detection routine were specifically written for the effort (P.B.) extending previous programs (9–11).
The output obtained (Fig. 2) allows rapid assignment of the different RNA features analyzed by simple visual inspection. Alternatively, results can be stored in an output file. The output gives first some general information on the RNA structure [energy, length, type (DNA/RNA), total stem-loop content] as well as motifs, stem-loops and structure templates in the different regions of an RNA (including 5′ UTR, CDS, 3′ UTR in mRNA) and retrieved specific RNA signals (polyA, protein A1 binding site, AU rich region, Sm-sites, poly A site motif, cleavage stimulation factor binding site) as well as broader motif classes ‘indication for processing protein binding motif’, ‘potential snRNP binding motif’.
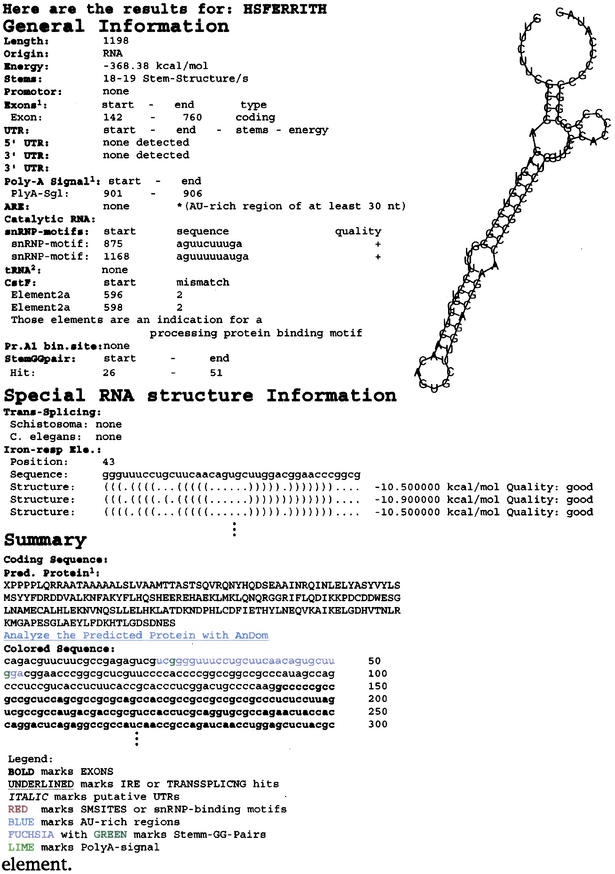
Figure 2.
An output example is given. Besides the specific regulatory element, an IRE in this example sequence, a number of further information items regarding regulatory elements are displayed. Note that only part of the sequence, of the folding and of the complete output is shown.
For RNAs below 1500 bp it shows the RNA folding and an overview on RNA stem loop content in the different regions. In addition, specific RNA stem-loop structures, trp-operon like structures, palindromes, G-G pairs (central regulatory feature in viral RNA elements such as the rev-response element), potential splice sites, pol III promotor binding sites are automatically displayed as well as potential sites for translational control, tRNA templates (subroutine using tRNA scan) and trans-splicing RNAs (specific template: catalytic leader). In addition, the text features colour-coded in the output (different regulatory motifs detected) are mapped on the proposed structure in its bracket notation to better identify structural regions of particular interest.
Further analysis results are summarized in Table 1. Any RNA can be submitted and analyzed by the server. Systematically general features are identified and summarized such as overall length, contained CDS etc. Furthermore, stem-loops, translational active short peptide regions and various protein binding sites are efficiently detected. The potential for the RNA to contain a specific structure is indicated, e.g. to be a structural RNA such as tRNA, to contain translational regulatory motifs and to be an RNA with catalytic potential. Furthermore, the RNA analyzer detects important specific functional RNAs for each of these categories of RNA elements: IREs, trans-splicing structures and tRNAs. In most browsers (e.g. Netscape) the obtained web server outputfile with the complete output can be exported or saved easily in html format or separately the picture can be saved as jpg and the text as a text file.
Efficiency
Response time of the server is fast: single sequences are analyzed within seconds, depending on the length and web traffic: on average, below 100 nt, 1 s calculation time is required; at 360 nt, 3 s; at 700 nt, 6 s; and at 1400 nt, 15 s calculation time. This included total folding of the molecule (<1500 basepairs) and detection of numerous sequence motifs, secondary structure features and marking the different regions of the RNA molecule (e.g mRNA regions). The server is implemented and ready to use at http://wb2x01.biozentrum.uni-wuerzburg.de/.
DISCUSSION
Server specific features
Our server allows a fast and comprehensive detection of RNA specific features in any nucleotide sequence. Each motif is analyzed directly and efficiently by subroutines following motif specific decision trees. They consider both primary sequence, secondary structure as well as simplified energy calculations. We provide in this way an integrated RNA analysis tool for detection of a couple of well-defined regulatory motifs and elements, allowing at the same time detection and analysis of primary sequence, secondary structure and specific folding. Discrete features on any of these levels are directly assigned to the user such as polyA site, stem-loop structure in translational region, trans-splicing structure or trans-splicing leader. In addition, there are alerts for potential structures. These are derived when the final structure was not found but several steps before pointed to a potential structure in this RNA region. Prompts such as ‘potential…motif’ alert the user that the regulatory motif scan by the server is no substitute for a specialist analysis including more programs and e.g. phylogenetic analysis of the RNAs in question. Thus Sm-sites detected are found by pattern matching to typical Sm-sites and should next be validated by further analysis. However, the tool is a handy primer for a first and swift systematic analysis regarding regulatory RNA motifs including built in specific identification of a couple of important regulatory RNA elements.
Main applications are quick analysis of specific RNAs, RNA families or genomic regions for functional features.
Several independent and different methods have been developed to analyze RNA structure and sequence features. The challenge in regulatory motif detection is the great complexity and variance of such motifs. For specific motifs, a number of approaches such as neural networks (14), hidden Markov models (4) and consensus structure approaches (15) have been developed. We decided a decision tree based approach to rapidly converge on specific motifs while keeping generality in the alerts for hidden structure, the overall folding picture and specific stem-loop scanning in different regions. We could achieve rapid response time. In this specific combination our tool adds to RNA analysis tools already available in the community. For instance, UTRscan (16) or ARE detection for the ARED database (17) exploit very efficiently pattern detection by versatile pattern search programs. However, folding is not considered. Rapid scanning of different RNAs including scanning their structure is offered by our tool-box, besides mfold-routines and fast scanning for stem-loops it offers search routines for specific RNA elements such as IRE and trans-splicing RNAs where it again rapidly scans for the structure as well as primary sequence features and includes a fast energy calculation. Our tool allows detection of 20 different regulatory features, including enhanced detection of certain highly specific regulatory elements. For example, besides the IREs in ferritin, ALAS and cell adhesion regulator 1 the recently experimentally confirmed IRE in NRAMP2 mRNA (18) is also detected by our approach (which is not detected by standard pattern matching using, for example, UTR-scan). This is nevertheless still only a selection focussing on our area of expertise. For instance, detection of SECIS (16) elements in the 3′ UTR is not yet included in the present version whereas for example automatic detection of CstF elements and G-G pairs are offered here for the first time to the community (Table 1). In addition our server considers also incomplete matches and scans as well as alerts the user for potential interesting RNA regions with a potential for a certain function (Table 1).
We have to stress, however, that the RNA analyzer scan is no substitute for a dedicated specialist analysis but rather a preparatory step for this, a first hand and easy to achieve overview. The present server gives a detailed overview on RNA specific features and regulatory motifs including structural information. It integrates a number of programs to delineate sequence features and motifs occuring in the RNA. Additional routines for further motif detection can be integrated for future development. Folding (<1500 nts) and detailed information on sequence specific structures as well as stem loop content in different regions is also supplied to better characterize functional features of the RNA. The program uses the mfold (5) routine of the Vienna package. To keep output small, only the top structure is displayed in our tool-box (if desired this can be changed in the source code). Detailed analysis of RNA folding such as looking for multiple foldings [program mfold, (5)] was not intended or attempted to keep rapid server response times. However, the summary information is already a first primer if one wants to hunt for a specific RNA structure. Thus the stem-loop scans in different regions can be analyzed further and it is of course possible to investigate detailed foldings in regions of interest. To rapidly test the reliability of the top structure displayed (according to the mfold routine, note that this is not too reliable for tRNA structures) the user simply includes modified versions of the original sequence. These can all be tested at the same time if submitted in fasta format and boxing the multiple sequence option in the server menu. With this option of the tool-box also regulatory elements or secondary structures conserved between different organisms or common among related RNAs can be rapidly scanned for. The program has been extensively tested for quick and reliable RNA analysis and swift identification of regulatory motifs by several bioinformatics courses as well as scientific users.
Acknowledgments
ACKNOWLEDGEMENT
We thank DFG for support (SFB 544/B2; BO-1099/5-2).
REFERENCES
- 1.Dandekar T. and Bengert,P. (2002) RNA Motifs and Regulatory Elements. 2nd Edn. Springer, New York. [Google Scholar]
- 2.Huynen M., Gutell,R. and Konings,D. (1997) Assessing the reliability of RNA folding using statistical mechanics. J. Mol. Biol., 267, 1104–1112. [DOI] [PubMed] [Google Scholar]
- 3.Eddy S.R. (2002) A memory-efficient dynamic programming algorithm for optimal alignment of a sequence to an RNA secondary structure. BMC Bioinformatics, 3, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eddy S.R. (1998) Profile hidden Markov models. Bioinformatics, 14, 755–763. [DOI] [PubMed] [Google Scholar]
- 5.Zuker M. (1994) Prediction of RNA secondary structure by energy minimization. Methods Mol. Biol., 25, 267–294. [DOI] [PubMed] [Google Scholar]
- 6.Zuker M. (2000) Calculating nucleic acid secondary structure. Curr. Opin. Struct. Biol., 10, 303–310. [DOI] [PubMed] [Google Scholar]
- 7.Hofacker I., Fontana,W., Stadler,P., Bonhoeffer,L., Tacker,M. and Schuster,P. (1994) Fast folding and comparison of RNA secondary structures (The Vienna RNA Package). Monatshefte fuer Chemie (Chemical Monthly), 125, 167–188. [Google Scholar]
- 8.Burge C. and Karlin,S. (1997) Prediction of complete gene structures in human genomic DNA. J. Mol. Biol., 268, 78–94. [DOI] [PubMed] [Google Scholar]
- 9.Dandekar T., Stripecke,R., Gray,N.K., Goossen,B., Constable,A., Johansson,H.E. and Hentze,M.W. (1991) Identification of a novel iron-responsive element in murine and human erythroid delta-aminolevulinic acid synthase mRNA. EMBO J., 10, 1903–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray N.K., Pantopoulos,K., Dandekar,T., Ackrell,B.A. and Hentze,M.W. (1996) Translational regulation of mammalian and Drosophila citric acid cycle enzymes via iron-responsive elements. Proc. Natl Acad. Sci. USA, 93, 4925–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dandekar T. and Sibbald,P.R. (1990) Trans-splicing of pre-mRNA is predicted to occur in a wide range of organisms including vertebrates. Nucleic Acids Res., 18, 4719–4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dandekar T. and Sharma,K. (1998) Regulatory RNA. Springer, Berlin; New York. [Google Scholar]
- 13.Lowe T.M. and Eddy,S.R. (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res., 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giddings M.C., Shah,A.A., Freier,S., Atkins,J.F., Gesteland,R.F. and Matveeva,O.V. (2002) Artificial neural network prediction of antisense oligodeoxynucleotide activity. Nucleic Acids Res., 30, 4295–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cannone J.J., Subramanian,S., Schnare,M.N., Collett,J.R., D'Souza,L.M., Du,Y., Feng,B., Lin,N., Madabusi,L.V. et al. (2002) The Comparative RNA Web (CRW) Site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics, 3, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pesole G. and Liuni,S. (1999) Internet resources for the functional analysis of 5′ and 3′ untranslated regions of eukaryotic mRNA. Trends Genet., 15, 378. [DOI] [PubMed] [Google Scholar]
- 17.Bakheet T., Williams,B.R. and Khabar,K.S. (2003) ARED 2.0: an update of AU-rich element mRNA database. Nucleic Acids Res., 31, 421–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tchernitchko D., Bourgeois,M., Martin,M.E. and Beaumont,C. (2002) Expression of the two mRNA isoforms of the iron transporter Nrmap2/DMTI in mice and function of the iron responsive element. Biochem. J., 363, 449–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt S., Bork,P. and Dandekar,T. (2002) A versatile structural domain analysis server using profile weight matrices. J. Chem. Inf. Comput. Sci., 42, 405–407. [DOI] [PubMed] [Google Scholar]