Abstract
Formation of numerous internal organs involves reciprocal epithelial–mesenchymal signaling and subsequent patterning and growth of the organ primordium. Capsulin is a basic helix–loop–helix transcription factor expressed in mesenchymal cells that encapsulate the epithelial primordia of internal organs, including the kidney and lung, as well as the epicardium, which gives rise to the coronary arteries. Capsulin is also expressed in the mesothelium that gives rise to the spleen. We demonstrate that mice homozygous for a capsulin null mutation fail to form a spleen. The homeobox genes Hox11 and Bapx1, shown previously to be essential regulators of spleen organogenesis, and a lacZ reporter introduced into the capsulin locus, were expressed in the early splenic primordium, derived from the splanchnic mesoderm, of homozygous mutant embryos. However, this primordium failed to develop beyond an initial group of precursor cells and underwent rapid apoptosis. The phenotype of capsulin mutant mice demonstrates that capsulin acts within a subpopulation of splanchnic mesodermal cells to control an essential early step in spleen organogenesis that is likely to represent a point of regulatory convergence of the capsulin, Hox11, and Bapx1 genes.
Organogenesis during vertebrate development requires cell fate specification and subsequent organization of precursor cells, often derived from multiple lineages, into precisely patterned tissues with highly specialized functions. Epithelial–mesenchymal interactions play essential roles in the development of a variety of internal organs, including lung, kidney, intestine, and pancreas (reviewed in ref. 1). Formation of these organs involves evagination of epithelial primordia at specific sites in response to signaling from adjacent mesenchyme. Reciprocal interactions between the coelomic epithelium of the dorsal mesogastrum and the underlying mesenchyme are also important for development of the spleen. Although the developmental importance of reciprocal signaling between epithelial and mesenchymal cells has been well documented, relatively little is known of the transcription factors that mediate these signaling events during organogenesis.
Members of the basic helix–loop–helix (bHLH) family of transcription factors have been shown to regulate development and differentiation of a wide range of cell types (reviewed in ref. 2). Capsulin (3, 4), also referred to as Pod-1 (5) and epicardin (6), is a bHLH transcription factor expressed in mesenchymal cells at sites of epithelial–mesenchymal interactions in the developing respiratory, gastrointestinal, urogenital, and cardiovascular systems, as well as in primordia of the spleen and in the epicardium, a mesenchymal cell layer that surrounds the heart and gives rise to the coronary arteries. The name, capsulin, is derived from its expression pattern in developing mesenchyme that “encapsulates” the epithelial primordia of internal organs (3).
Capsulin binds the E-box consensus sequence (CANNTG) as a heterodimer with the ubiquitous bHLH protein E12, but it lacks a transcription activation domain (3). The bHLH region of capsulin is nearly identical to that of MyoR, which is expressed in undifferentiated skeletal myoblasts in culture and early in the skeletal muscle lineage in vivo (7, 8). MyoR acts as a potent transcriptional repressor that can block myoblast differentiation by interfering with the activity of MyoD (7). The functions of capsulin and MyoR in vivo remain to be determined, but their sequence homology, abilities to bind the same DNA sequence as heterodimers with E12, and lack of transcriptional activity suggest that these bHLH proteins play similar roles in the lineages in which they are expressed.
In the present study, we investigated the function of capsulin during mouse embryogenesis by creating capsulin mutant mice. The phenotype of homozygous capsulin mutants reveals a critical role for capsulin in the formation of the spleen. Capsulin acts after splenic specification to control morphogenetic expansion of the splenic anlage and in its absence, splenic precursor cells undergo programmed cell death. This splenic phenotype, which resembles that of mice lacking the homeobox genes Hox11 (9, 10) and Bapx1 (11, 12), suggests that capsulin, Hox11, and Bapx1 may control a common essential early step in the developmental pathway for spleen organogenesis.
Methods
Gene Targeting and Creation of Mutant Mice.
Capsulin targeting vectors were created from capsulin genomic clones isolated from a 129svEv mouse genomic library. The capsulin gene contains two exons separated by a 1.7-kb intron. Exon 1 encompasses the coding sequence for amino acids 1–150, including the bHLH region. Two different targeting constructs were created. In one construct, all coding sequence from exon 1 was replaced with a PGKneo cassette, to confer neomycin resistance. The 5′ arm of homology was obtained by PCR from the genomic clone and was cloned upstream of PGKneo. A thymidine kinase (tk) gene under the control of the herpes simplex virus promoter was linked to the 3′ arm of homology. In the other construct, the 5′ arm of homology was subcloned upstream of a promoterless β-galactosidase gene. This capsulin-lacZ cassette was then subcloned upstream of PGKneo. This targeting construct had the same 3′ arm of homology and tk cassette as the former construct.
The linearized targeting vectors were electroporated into 129 embryonic stem (ES) cells, which were then plated onto G-418-resistant mitotically inactivated STO fibroblasts. ES cell clones were isolated after positive and negative selection with G-418 (Geneticin, 180 μg/ml of active concentration, GIBCO/BRL) and 0.2 μM FIAU [1,2-deoxy-2-fluoro-β-d-arabinofuranosyl)-S-iodouracil], respectively, as described (13). Site-specific recombination events were identified by Southern blot analysis with SacI, which distinguished the wild-type and targeted alleles because of introduction of a SacI site into the capsulin locus by the targeting vector. SacI digestion and hybridization with a probe from outside the 5′ end of the targeting vector (shown in Fig. 1) resulted in a wild-type band of 6.0 kb and a mutant band of 3.5 kb.
Figure 1.
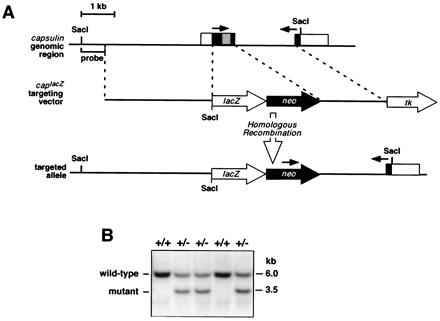
Targeting the capsulin locus. (A) The structure of the capsulin gene and surrounding genomic region is shown at the top, the lacZ targeting vector is in the middle, and the targeted allele is at the bottom. Noncoding region is in white, coding region in black, and the bHLH region in gray. The targeting vector introduced lacZ in-frame with the initiation codon of capsulin and a neomycin-resistance gene immediately downstream. The thymidine kinase gene at the 3′ end of the targeting vector was used for negative selection. The targeting strategy resulted in deletion of the entire first exon, which encodes the bHLH region. The position of the external probe used for Southern analysis in B is indicated. Genotyping of ES cells and offspring was performed by SacI digestion of DNA and hybridization to the indicated probe or by PCR. Arrows designate the positions of PCR primers used for genotyping. (B) Southern blot of genomic DNA from wild-type (+/+) and capsulin+/− mice. The probe shown in A, which is external to the region of vector homology, detects a wild-type band of 6.0 kb and a mutant band of 3.5 kb, after digestion with SacI.
From over 3,000 independent ES cell clones analyzed, the targeting frequency of both vectors was about 1:400. ES cells heterozygous for the targeted capsulin allele were injected into C57BL/6 blastocysts, which were implanted into pseudopregnant Swiss foster female mice to obtain chimeric offspring. Both mutations were transmitted through the germline, as confirmed by Southern analysis of tail genomic DNA, as described above. Mice heterozygous for the capsulin mutations were phenotypically normal and were intercrossed to obtain homozygous mutants. Mice were produced in the Black Swiss × 129/SvEv mixed genetic background.
Histological Analysis and lacZ Staining.
β-Galactosidase staining was performed as described (14). Embryos and tissues were prepared for sectioning by dehydration in ethanol and embedding in paraffin. Sections were cut at 10-μm intervals and dried on microscope slides. Paraffin was removed with xylene, and sections were stained with hematoxylin/eosin or simply treated with cytoseal and covered.
Terminal deoxynucleotidyltransferase-mediated UTP end labeling (TUNEL) staining for apoptotic cells was done on sections from paraformaldehyde-fixed embryos embedded in paraffin according to the protocol supplied with Promega Apoptosis Detection System. Apoptotic cells were labeled with fluorescein, and sections were counterstained with propidium iodide.
In Situ Hybridization.
Radioactive in situ hybridization was performed on paraffin-embedded sections of embryos by using 35S-radiolabeled RNA probes for Hox11 (9) and Bapx1 (11), as described (15).
Results
Generation of capsulin Mutant Mice.
Two different targeting vectors were used to inactivate the mouse capsulin gene (see Materials and Methods); both replaced the first exon, encoding the bHLH region, with a neomycin-resistance cassette, resulting in null mutations, and one introduced a lacZ gene in-frame with the initiation codon (Fig. 1A). Multiple independent ES cell clones harboring the targeted alleles were injected into C57BL/6 blastocysts to obtain chimeric offspring that transmitted the mutations through the germline. Mice heterozygous for the capsulin null allele appeared phenotypically normal and were bred to obtain homozygous mutants. Offspring from capsulin+/− intercrosses were genotyped by Southern analysis of tail genomic DNA (Fig. 1B). Capsulin heterozygous and homozygous mutants were obtained at predicted Mendelian ratios (data not shown).
Failure in Spleen Organogenesis in capsulin Mutant Mice.
Capsulin−/− offspring were born alive and appeared grossly normal. However, they exhibited gasping motions because of difficulty in breathing, quickly became pale and cyanotic, and died within 5–10 min after birth. As described previously (16), the lungs and kidneys of homozygous mutants were severely hypoplastic (data not shown). Failure of the lung epithelia to undergo branching morphogenesis and the resulting absence of alveoli likely account for early perinatal lethality of the mutant. In addition, gross anatomical analysis of internal organs of mutant neonates revealed the complete absence of a spleen (Fig. 2), which was not observed in a previous study of capsulin mutant mice (16). The stomach and pancreas, which are derived from a region of splanchnic mesoderm adjacent to the splenic anlage (17), were properly located in the mutant (Fig. 2). The phenotypes resulting from our two different capsulin mutations were identical. Although capsulin is also expressed in myogenic precursor cells within the branchial arches and in the epicardium (Fig. 3 and refs. 3–6), which gives rise to the coronary arteries, we did not detect defects in these tissues of capsulin mutants.
Figure 2.
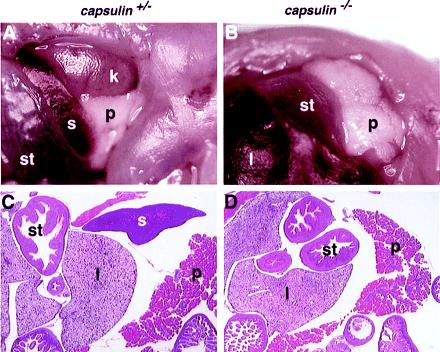
Asplenia of capsulin mutant mice. (A and B) The abdomen of capsulin+/− and capsulin−/− neonates was dissected to reveal the internal organs. The spleen(s) in the homozygous mutant (B) is completely missing, but the other internal organs are present. Because the kidney (k) is underdeveloped in the mutant, it is covered by the pancreas (p) and is not observable from this view. l, liver; st, stomach. (C and D) Capsulin+/− and capsulin−/− neonates were sectioned, and internal organs were analyzed after hematoxylin/eosin staining. The spleen is readily detectable in the heterozygous animal but not in the homozygous mutant.
Figure 3.
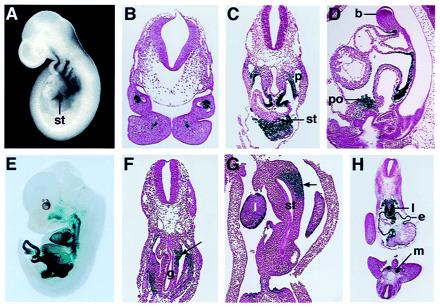
Embryonic expression pattern of lacZ from targeted capsulin allele. Embryos homozygous for the capsulin-lacZ knockin allele were stained for lacZ expression, sectioned, and stained with hematoxylin/eosin for histologic analysis. A and E show whole-mount embryos at E9.5 and E13.5, respectively. The vertical line in A points to the septum transversum (st). (B) Transverse section through the branchial arches at E9.5 showing a core of lacZ-positive cells, representing skeletal muscle progenitors. (C) Transverse section through an E10.5 embryo at the level of the posterior region of the heart. LacZ staining can be seen in the region of the septum transversum (st) below the heart and in the mesothelium along the pleural cavity (p), adjacent to the future lung buds. (D) Sagittal section through an E10.5 embryo showing lacZ staining within the proepicardial organ (po) and epicardium. b, branchial arch. (F) Transverse section through an E10.5 embryo more posterior than in C, showing asymmetric lacZ staining in visceral mesothelium and adjacent mesoderm on the left side of the embryo and adjacent to the gut (g), representing the splenic anlage (indicated by arrow). (G) Transverse section at E11.5 through the dorsolateral region of the stomach (st) and splenic primordium, showing asymmetric left-sided expression of lacZ (indicated by arrow). l, liver. (H) Transverse section at E10.5 showing lacZ staining in the lung primordium (l), epicardium (e), and condensing metanephric mesenchyme (m).
Expression of lacZ from the capsulin Locus.
By staining for lacZ, we were able to correlate sites of capsulin gene expression in wild-type and mutant offspring with possible developmental abnormalities in the mutants and identify embryonic sites of capsulin expression with greater precision than from in situ hybridization. LacZ expression from the mutant capsulin allele mirrored expression of the endogenous gene throughout embryogenesis (Fig. 3 and data not shown). LacZ expression was first detected at embryonic day (E)8.5 in mesodermal cells migrating into the first and second branchial arches (data not shown). Between E9.5 and E13.5, the pattern of lacZ expression was indistinguishable in capsulin+/− and capsulin−/− embryos, except in the splenic primordium (see below).
At E9.5, lacZ staining was observed in skeletal muscle precursors in the branchial arches and in a grape-like cluster of cells, known as the proepicardial organ, located in the region of the septum transversum immediately posterior to the heart (Fig. 3A). These mesothelial cells migrate anteriorly over the heart, giving rise to the epicardium, the source of progenitors for smooth muscle and endothelial cells of the coronary vessels (18). Within the branchial arches, lacZ expression was localized to a core of mesenchymal cells, adjacent to the branchial arch arteries (Fig. 3B). These cells appear to correspond to skeletal muscle precursors derived from the paraxial mesoderm, which gives rise to all voluntary facial muscles arising from the branchial arches (19).
At E10.5, lacZ expression was observed in a bilaterally symmetrical pattern in the mesothelium lining the visceral and parietal aspects of the pericardio–peritoneal canals (Fig. 3C). Strong expression was also present in the hepatic primordium within the septum transversum. LacZ expression in the proepicardial organ and epicardium was readily apparent in sagittal sections at E10.5 (Fig. 3D). Transverse sections at this stage also showed lacZ staining in the epicardium, as well as in the condensing metanephric mesenchyme and lung primordium (Fig. 3H).
In transverse sections through the region where the foregut expands to form the stomach, lacZ expression was asymmetrical at E10.5, appearing only in the visceral mesothelium and adjacent mesoderm on the left side of the embryo (Fig. 3F). At E11.5, lacZ expression was also evident within a focal region of undifferentiated mesoderm corresponding to the embryonic origin of the spleen in the dorso-lateral wall of the stomach (Fig. 3G). We interpret the presence of these early lacZ-positive mesenchymal cells in capsulin −/− embryos as an indication that capsulin is not required for specification of the splenic precursor cell population.
The spleen forms from an amalgamation of several aggregates of mesodermal tissue within the dorsal mesentery. Spleen development begins at about E11.5 in the mouse, as splanchnic mesodermal cells condense around the dorsal mesogastrum, a specific region of thickened epithelium along the dorsolateral region of the future stomach (9, 17). The splenic primordium first becomes morphologically identifiable as a ridge of cells immediately dorsal to the stomach at about E13.5. Capsulin is expressed in splanchnic mesodermal cells immediately adjacent to the coelomic epithelium at E11.5–12.5, when spleen development commences (3).
At E13.5, lacZ staining was apparent in the developing spleen of capsulin+/− embryos (Fig. 4A). However, in capsulin−/− embryos, there was only a faint trace of lacZ-positive cells in an acellular region where the spleen should have formed (Fig. 4B). At E13.5 (Fig. 3E) and later (data not shown), lacZ expression was observed in the jaw muscles and tongue, consistent with the earlier expression in myogenic precursors within the branchial arches, as well as in the epicardium and smooth muscle layers of the esophagus and gastrointestinal and urogenital tracts of capsulin+/− and capsulin−/− mice.
Figure 4.
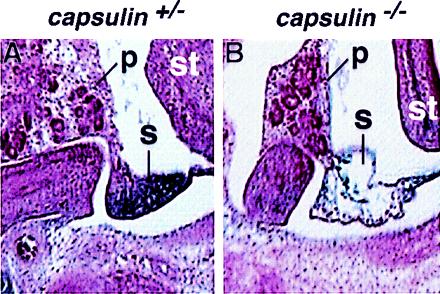
Histologic analysis of the developing spleen at E13.5. A shows a section through the developing spleen and surrounding tissues of a capsulin+/− embryo at E13.5. LacZ staining is apparent in the spleen. B shows a section through a capsulin−/− embryo at the same level as in A. Note the region of the developing spleen is virtually devoid of cells, whereas the adjacent pancreas and stomach appear normal. p, pancreas; s, spleen, st, stomach.
Spleen Specification but Not Organogenesis in capsulin Mutant Mice.
To determine whether other early markers of spleen development were expressed in capsulin−/− embryos, we examined, by in situ hybridization, expression of the homeobox genes Hox11 and Bapx1, both of which are coexpressed with capsulin in the developing spleen and are required for spleen development (9–12). Transcripts for Hox11 and Bapx11 were readily detected in the developing spleen of wild-type embryos from E12.5–14.5 (Fig. 5). In mutant embryos, we detected Hox11 and Bapx11 transcripts in the region of the splenic primordium at E12.5, consistent with the conclusion that splenic precursors are specified in the mutant, but thereafter this region did not expand to give rise to the morphologically recognizable organ, and all traces of expression of these markers disappeared from the presumptive splenic primordium (Fig. 5).
Figure 5.
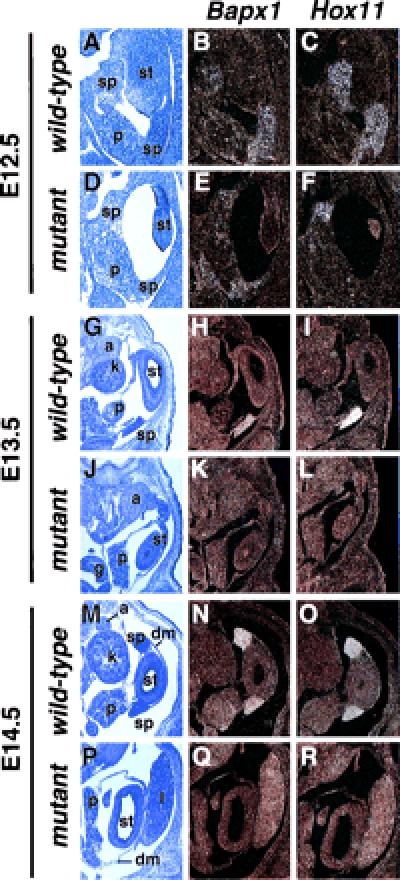
Absence of Hox11 and Bapx1 expression in splenic primordium of capsulin−/− mice. Bapx1 and Hox11 transcripts were detected by in situ hybridization to abdominal sections of wild-type and capsulin mutant embryos at E12.5–14.5, as indicated. Both transcripts were detected in the region of the splenic anlage at E12.5 but were not detected thereafter. The sections on the left are representative sections of adjacent wild-type or mutant embryos stained with hematoxylin/eosin. a, adrenal gland; dm, dorsal mesogastrum; g, gut; k, kidney; l, liver; p, pancreas; sp, spleen; st, stomach.
Apoptosis of the Splenic Primordium in capsulin Mutant Mice.
In light of the apparent absence of splenic cells in capsulin mutant embryos after E12.5, we performed TUNEL staining on histological sections to determine whether there was an increase in apoptotic cell death in the splenic lineage of the mutants. Indeed, as shown in Fig. 6, apoptosis was readily detected in the presumptive splenic-forming region of mutant embryos, whereas only random and occasional TUNEL staining was observed in the internal organs of wild-type embryos, without preferential localization to any particular tissue.
Figure 6.
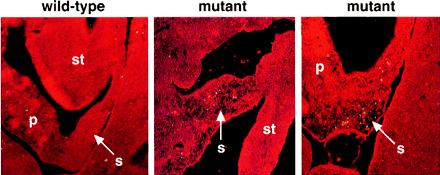
Apoptosis in the splenic lineage of capsulin mutant embryos revealed by TUNEL staining. Histologic cross sections through abdominal sections of wild-type and capsulin mutant embryos at E12.5 were stained by TUNEL staining to detect apoptosis. Center and Right show the dorsal and ventral regions, respectively, of the developing spleen with extensive apoptosis (yellow) localized to the splenic primordium. p, pancreas; s, spleen; st, stomach. In the wild-type embryo (Left), there is only an occasional TUNEL-positive cell with no preferential localization to any particular tissue. The wild-type section corresponds to an expanded region of the section in Fig. 5B, and the mutant sections correspond to expanded regions of the section in Fig. 5C.
Discussion
The phenotype of capsulin mutant mice reveals an essential role for capsulin in organogenesis of the spleen. Based on expression of Hox11 and Bapx1, as well as lacZ from the targeted capsulin allele, we conclude that capsulin acts after the initial specification step for the splenic lineage and is required for expansion of the splenic anlage. In the absence of capsulin, splenic precursors undergo apoptotic cell death. Capsulin mutant mice also exhibit severe defects in kidney and lung morphogenesis that include abnormalities in epithelial branching morphogenesis that result in hypoplastic atrophic kidney glomeruli and a lack of lung alveoli (ref. 16 and data not shown).
Formation of the spleen, lung, and kidney requires reciprocal interactions between mesenchymal precursors and adjacent epithelial primordia (1). Although the developmental defect in the spleen is much more severe than the lung and kidney defects, it is possible that capsulin regulates a step in epithelial–mesenchymal interactions common to each of these developing organs. In the developing lung, capsulin has been shown to be required in the mesenchyme to activate expression of bone morphogenetic protein-4 (BMP-4) in the adjacent epithelium; in the absence of BMP-4 expression, the airway epithelium fails to differentiate (16). BMP-4 is also expressed in the visceral mesoderm during gut development (20), making it a potential mediator of capsulin actions in spleen development, although we are unaware of direct evidence for a role of BMP-4 signaling in spleen development per se.
The asplenic phenotype of capsulin mutant mice resembles that of mice lacking Hox11 (9, 10) and Bapx1 (11, 12), as well as the Wilm's tumor suppressor gene, WT-1 (21), which are coexpressed with capsulin during spleen development, leading us to conclude that these genes may act in concert to regulate a common early event in spleen organogenesis. Mice lacking the homeobox gene Nkx2–3 also show abnormalities in spleen development with varying penetrance (22). In this regard, recent studies have demonstrated specific interactions between bHLH and homeodomain proteins involved in pituitary development (23), and we have found that capsulin interacts directly with Bapx1, although the physiological significance of this interaction remains to be determined (J.L. and E.N.O., unpublished results). Interestingly, Hox11 is also coexpressed with capsulin in the muscle plates of the branchial arches and the epicardium of the heart (Fig. 3 and refs. 3–6), suggesting that these transcription factors act within a common developmental pathway in these tissues.
Within the bHLH region, capsulin shares high homology with MyoR, a transcriptional repressor expressed in the early skeletal muscle lineage (7). Capsulin also can act as a transcriptional repressor (J.L. and E.N.O., unpublished results). How might a transcriptional repressor regulate an early step in organogenesis? One possibility is that capsulin might normally suppress differentiation, thereby allowing a precursor cell pool to expand to generate the organ, or it might repress expression of a repressor of organ development. It is also possible that capsulin may act as a repressor or an activator of transcription, depending on cell type or cell environment.
The expression of capsulin and MyoR overlaps in head muscle precursors, raising the possibility that the lack of a phenotype in the head musculature of capsulin mutant mice may reflect functional redundancy between capsulin and MyoR. Mice homozygous for a MyoR null mutation are viable (J.L. and E.N.O., unpublished data), which will make phenotypic analysis of capsulin/MyoR double mutants particularly interesting. Notably, the Drosophila bHLH factor bHLH54 is closely related to capsulin and MyoR and appears to exhibit their combined expression patterns, being expressed specifically in visceral and a subset of skeletal muscle precursor cells during embryogenesis (24). Visceral muscle development in Drosophila is controlled by the homeobox gene bagpipe (25), an ortholog of Bapx1. Thus, the requirement of capsulin and Bapx1 for spleen development raises the intriguing possibility that bHLH (capsulin/bHLH54F) and homeobox (Bapx1/bagpipe) genes may act within evolutionarily conserved genetic pathways for organogenesis. Identifying the target genes for these transcription factors and determining the potential significance of the interactions between the factors represent important issues for the future.
Acknowledgments
We are grateful to T. Lufkin and S. Korsmeyer for providing reagents, S. Comerfeld for assistance with analysis of apoptosis, and R. MacDonald for comments on the manuscript. We thank J. Page for editorial assistance and A. Tizenor for graphics. This work was supported by grants from the National Institutes of Health, the D. W. Reynolds Foundation, and the American Heart Association to E.N.O.
Abbreviations
- bHLH
basic helix–loop–helix
- ES
embryonic stem
- En
embryonic day n
- TUNEL
terminal deoxynucleotidyltransferase-mediated UTP end labeling
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on May 2, 2000
References
- 1.Thesleff I, Vaahtokari A, Partanen A M. Int J Dev Biol. 1995;39:35–50. [PubMed] [Google Scholar]
- 2.Massari M E, Murre C. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu J, Richardson J A, Olson E N. Mech Dev. 1998;73:23–32. doi: 10.1016/s0925-4773(98)00030-6. [DOI] [PubMed] [Google Scholar]
- 4.Hidai H, Bardales R, Goodwin R, Quertermous T, Quertermous E E. Mech Dev. 1998;73:33–43. doi: 10.1016/s0925-4773(98)00031-8. [DOI] [PubMed] [Google Scholar]
- 5.Quaggin S E, Vanden Heuvel G B, Igarashi P. Mech Dev. 1998;71:37–48. doi: 10.1016/s0925-4773(97)00201-3. [DOI] [PubMed] [Google Scholar]
- 6.Robb L, Mifsud L, Hartley L, Biben C, Copeland N G, Gilbert D J, Jenkins N A, Harvey R P. Dev Dyn. 1998;213:105–113. doi: 10.1002/(SICI)1097-0177(199809)213:1<105::AID-AJA10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, Webb R, Richardson J A, Olson E N. Proc Natl Acad Sci USA. 1999;96:552–557. doi: 10.1073/pnas.96.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robb L, Hartley L, Wang C C, Harvey R P, Begley C G. Mech Dev. 1998;76:197–201. doi: 10.1016/s0925-4773(98)00122-1. [DOI] [PubMed] [Google Scholar]
- 9.Roberts C W-M, Shutter J R, Korsmeyer S J. Nature (London) 1994;368:747–749. doi: 10.1038/368747a0. [DOI] [PubMed] [Google Scholar]
- 10.Dear T N, Colledge W H, Carlton M B L, Lavenir I, Larson T, Smith A J H, Warren A J, Evans M J, Sofroniew M V, Rabbitts T H. Development (Cambridge, UK) 1995;121:2909–2915. doi: 10.1242/dev.121.9.2909. [DOI] [PubMed] [Google Scholar]
- 11.Tribioli C, Lufkin T. Development (Cambridge, UK) 1999;126:5699–5711. doi: 10.1242/dev.126.24.5699. [DOI] [PubMed] [Google Scholar]
- 12.Lettice L A, Purdie L A, Carlson G J, Kilanowski F, Dorin J, Hill R E. Proc Natl Acad Sci USA. 1999;96:9695–9700. doi: 10.1073/pnas.96.17.9695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin J F, Bradley A, Olson E N. Genes Dev. 1995;9:1237–1249. doi: 10.1101/gad.9.10.1237. [DOI] [PubMed] [Google Scholar]
- 14.Cheng T-C, Wallace M, Merlie J P, Olson E N. Science. 1993;261:215–218. doi: 10.1126/science.8392225. [DOI] [PubMed] [Google Scholar]
- 15.Shelton J M, Lee M-H, Richardson J A, Patel S B. Lipid Res. 2000;41:532–537. [PubMed] [Google Scholar]
- 16.Quaggin S E, Schwartz L, Cui S, Igarashi P, Deimling J, Post M, Rossant J. Development (Cambridge, UK) 1999;126:5771–5783. doi: 10.1242/dev.126.24.5771. [DOI] [PubMed] [Google Scholar]
- 17.Green M C. Dev Biol. 1967;15:62–89. doi: 10.1016/0012-1606(67)90006-1. [DOI] [PubMed] [Google Scholar]
- 18.Virace S, Challice C E. Anat Rec. 1981;201:157–168. doi: 10.1002/ar.1092010117. [DOI] [PubMed] [Google Scholar]
- 19.Noden D M. J Anat. 1993;186:257–276. [Google Scholar]
- 20.Roberts D J, Johnson R L, Burke A C, Nelson C E, Morgan B A, Tabin C. Development (Cambridge, UK) 1995;121:3163–3174. doi: 10.1242/dev.121.10.3163. [DOI] [PubMed] [Google Scholar]
- 21.Herzer U, Crocoll A, Barton D, Howells N, Englert C. Curr Biol. 1999;9:837–840. doi: 10.1016/s0960-9822(99)80369-8. [DOI] [PubMed] [Google Scholar]
- 22.Pabst O, Zwelgerdt R, Arnold H-H. Development (Cambridge, UK) 1999;126:2215–2225. doi: 10.1242/dev.126.10.2215. [DOI] [PubMed] [Google Scholar]
- 23.Poulin G, Lebel M, Chamberland M, Paradis F W, Drouin J. Mol Cell Biol. 2000;20:4826–4837. doi: 10.1128/mcb.20.13.4826-4837.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Georgias C, Wasser M, Hinz U. Mech Dev. 1997;69:115–124. doi: 10.1016/s0925-4773(97)00169-x. [DOI] [PubMed] [Google Scholar]
- 25.Azpiazu N, Frasch M. Genes Dev. 1993;7:1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]


