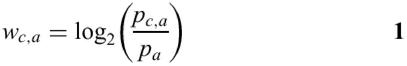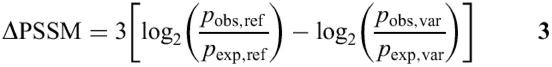Abstract
PARSESNP is a tool for the display and analysis of polymorphisms in genes. Using a reference DNA sequence, an exon/intron position model and a list of polymorphisms, it determines the effects of these polymorphisms on the expressed gene product, as well as the changes in restriction enzyme recognition sites. It shows the locations and effects of the polymorphisms in summary on a stylized graphic and in detail on a display of the protein sequence aligned with the DNA sequence. The addition of a homology model, in the form of an alignment of related protein sequences, allows for prediction of the severity of missense changes. PARSESNP is available on the World Wide Web at http://www.proweb.org/parsesnp/.
INTRODUCTION
There is a plethora of information about sequence variants available on the World Wide Web, ranging from medical data at the Human Gene Mutation Database (1) (http://www.hgmd.org/) and dbSNP (2) (http://www.ncbi.nlm.nih.gov/SNP/) to the results of reverse genetic screens in plants (3) (http://tilling.fhcrc.org:9366/). A problem that comes up frequently when attempting to work with this data is how to visualize the effects of the variants. We have developed PARSESNP (Project Aligned Related Sequences and Evaluate SNPs), a web-based tool which integrates information from these various sources of polymorphism data and provides a consistent, user-friendly interface for the analysis of genetic polymorphisms.
The tool has been designed with flexibility in mind: it can use polymorphism data (in either DNA sequence or translated protein sequence) from a number of databases, it can compare these variants to genomic or cDNA reference sequence and it can process small insertions and deletions in addition to SNPs. The output of the program provides information that is useful to many different applications, including predictions of the severity of missense changes and an indication of the restriction enzyme polymorphisms that result from a change. It has been used as a data analysis tool in both forward genetics (4) and in reverse genetics (5).
SOURCES OF MUTATIONS AND VARIANTS
The polymorphisms can be entered automatically from a variety of sources, including the HGMD, dbSNP, GenBank (2) (http://ncbi.nlm.nih.gov) and SWISS-PROT (6) (http://www.expasy.ch/) on-line databases. Users can supply their own polymorphisms, either through a web form or an uploaded file, as a nucleotide change, expressed either as an absolute position or a modified fragment or as an amino acid change in translated product. Other databases, such as the Arabidopsis TILLING Project, can pass their data directly to PARSESNP through the web interface, allowing for the seamless integration of separate applications. Despite the limitation its name implies, the current version of PARSESNP is capable of handling changes to multiple nucleotides, including small insertions and deletions.
Variants are determined relative to the supplied reference DNA sequence and gene model. These variants can be recognized in a number of formats.
Nucleotide change and position relative to the start of the DNA sequence, e.g. A123G.
Protein change and position relative to the start of gene product, e.g. M17V.
String of DNA with one change relative to reference sequence, e.g. ATGATGATG G TGATGATG.
Explicit change in DNA, e.g. ATGATGATG[A/G]TGATG.
Implicit change in DNA using the standard IUPAC codes, e.g. ATGATGATG R TGATG.
Insertions, e.g. A123AA or ATGATGATG[A/AA]TGATG and deletions, e.g. A123: or ATGATGATG[A/-]TGATG.
When the position of the change in the variant sequence and the location of the variant sequence on the reference sequence are unknown (format 3 above), they are determined using simple substring matching. When the position of the change in the variant sequence is known (formats 4, 5 and the second example in 6), all matching of variant sequence to reference sequence is done using BLAST (7) to allow imperfect matching in the region surrounding the change. The underlying codon change that results in an amino acid change (format 2) is determined using brute force, by examining all possible single nucleotide changes. If more than one nucleotide change could cause the specified change in protein sequence, each change is listed separately. In all cases, the original nucleotide or residue from the variant is checked against the reference sequence to verify that the variant corresponds to a valid change in the reference sequence; this is useful given the inconsistencies in nucleotide and residue numbering present in databases.
Multiple changes in the same individual can be grouped together to create more complicated patterns; for example, the change of a codon from ATG to TAG could be entered as either …[AT/TA]… or A1T,T2A (or even A1:,T2TA). Changes entered together in this way will be considered grouped for the purpose of determining restriction enzyme polymorphisms; however, each modified codon is considered separately for the purpose of determining its effect on protein function.
EFFECTS OF NUCLEOTIDE CHANGES
Effects on the gene
Effects on translation can be determined fairly easily from the gene model. PARSESNP detects truncation changes (both as changes to a stop codon in coding sequence and as changes in a splice junction at the beginning or end of an intron), missense changes and silent changes (in both coding and noncoding sequence). For insertions and deletions, it reports whether the polymorphism causes a frameshift.
In addition to direct effects on translation, PARSESNP determines the restriction enzyme sites present on the gene both before and after the introduction of the polymorphisms, using enzymes present in the REBASE database (8). It provides hyperlinks to the REBASE entries indicating the specific restriction enzyme sites gained in or lost from the reference sequence. This information can be helpful in testing biological samples for the presence of the polymorphism. As mentioned above, multiple changes combined to describe a more complex variant are considered together when determining restriction enzyme polymorphisms, so it is important to remember that each polymorphism entered should correspond to the combination of changes present in an individual.
Homology models
In order to assess the effect missense changes have on gene product function, PARSESNP provides two different and complementary methods of submitting homology information. A user can provide an alignment of protein sequences in any of several popular formats, including FASTA and ClustalX. In addition, PARSESNP accepts Blocks, a format that represents distinct regions of ungapped alignment in protein sequences, both from the Blocks database (9) and created by a user using the Blockmaker program (10); more information about Blocks is available at http://blocks.fhcrc.org/.
Sequence alignments are converted to Blocks format using programs available as part of the BLIMPS package (11). The blocks are then converted into a Position-Specific Scoring Matrix or PSSM, using another program (12) available as part of BLIMPS. In general, a PSSM gives a score for each amino acid at each position in a block; amino acids more represented at a position will be given a higher score at that position, while those less represented at that position will be given a lower score. The PSSM score wc,a for column c of the alignment and amino acid a is expressed in terms of the observed frequency pc,a of amino acid a in column c and the expected frequency pa of amino acid a.
Such scores range over the real numbers. Some information from a substitution matrix is used to compensate pc,a for amino acids unrepresented or underrepresented in the aligned column. This type of score is commonly referred to as a log-odds score; it is scaled by a factor for three to be consistent with other scoring matrices. The PSSM is then aligned to the gene using MAST (13), the Multiple Alignment Search Tool. This determines the mapping of the PSSM onto the sequence that maximizes the overall score for the sequence (from summing the scores in the columns of the PSSM corresponding to the amino acids found in the sequence). Matches are however limited to those where the probability (or p-value) of finding a match to the block as good as that found on the reference sequence in a random protein sequence is <10−4; this removes from consideration many undesirable, weak matches. Some blocks may be found more than once on the sequence, others may not be found at all or they may be found out of order. While this positional information may give an indication to the user of the quality of the homology model, PARSESNP accepts all block matches with a low enough p-value.
Effects on gene product function
The severity of the effect of a missense change on function can be predicted using the homology models. If the region containing a missense change is aligned to a block, one can attempt to gauge the effect of the change by examining the change in the PSSM score w that the variant would cause; we define this difference score as
This can be an informative measure, since some changes can actually bring the protein sequence closer to the alignment (indicated by a negative ΔPSSM) and are unlikely to have interesting effects, while others may move the protein sequence dramatically away from the alignment (indicated by a large positive ΔPSSM). It is, of course, impossible to choose a single value as a cutoff for a deleterious change. We have chosen 10 as a rough lower bound for the scores of changes predicted to be deleterious. Substituting Equation 1 into Equation 2 and including the scaling, we derive
from which it follows that a ΔPSSM score of 10 corresponds to a decrease of ∼10 in the odds-ratio ([( pobs,ref /pobs,var)/( pexp,ref /pexp,var)] = 210/3 ≈ 10.07) or, put another way, that the variant amino acid is 10-fold less likely to appear in that position of the alignment after correcting for overall amino acid frequencies. We find that results using this cutoff correlate well with those from the other prediction method discussed below.
If an alignment containing the query sequence is submitted, we additionally attempt to determine the severity of a missense change using the SIFT program (14), which assigns a score between 0 and 1 to each change from the reference sequence, with lower scores indicating a more deleterious change. SIFT scores <0.05 have been empirically determined to be deleterious. As mentioned above, this gives similar results to a ΔPSSM cutoff of 10 (for examples of this, see 4).
The quality of the predictions made can be no better than the quality of the alignment submitted; the old adage ‘garbage in, garbage out’ is as applicable here as anywhere else. The protein sequences in the homology model must be sufficiently diverged as to include alignment positions that have changed over time without loss of function, but not so broad as to include sequences that have lost or changed function. A good way to measure this is the median information content (15,16) of each block, which is displayed on the output; it ranges from 0.0 to 4.32 (log220, where 20 is the number of amino acids) and a reasonably diverged block has an information content of ∼3.
In addition to the quality of the alignment, the quality of its match to the reference sequence can have a profound effect on the accuracy of predictions. PARSESNP reports the p-value for each block hit on the output; values <10−10 indicate a strong match. Each residue on the reference sequence is colored green, black or red depending on how well that reference residue matches the aligned block; green indicates a PSSM score >0, red indicates a PSSM score <−2, and black indicates an intermediate score. Since the ΔPSSM score is more likely to be low at a reference residue that matches the block poorly, this can provide an additional means to evaluate extremely low ΔPSSM scores. Finally, one can also consider the order in which blocks match the sequence; a family in which all of the blocks match the query sequence in order is likely a better homology model for that sequence than one in which only a subset of the blocks match or some match out of order.
OUTPUT FORMATS
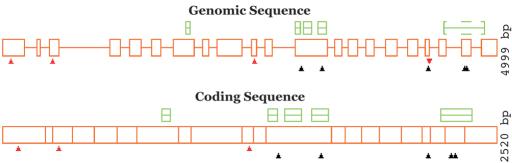
The results of PARSESNP can be viewed in a variety of different formats. At the top of the output page is a graphical plot of the locations of the polymorphisms on the gene (both coding sequence and genomic sequence, when applicable), along with indications as to the effects of each polymorphism and the locations of blocks on the sequence (Fig. 1). This is followed by a table (shown in Fig. 2) describing in detail the effect of each polymorphism on the gene, including nucleotide change, amino acid effect, restriction enzyme polymorphisms and ΔPSSM. Following the table is a detailed view of the reference sequence (and the spliced coding sequence when genomic sequence is provided) showing the DNA sequence, the corresponding amino acids, the polymorphisms and their effects and the block hits (Fig. 3). Finally, PARSESNP provides a link to the 3D Blocks program (9) which can display the positions of variants found in conserved regions on the structure of a similar protein, if one is available.
Figure 1.
Overview of polymorphisms and blocks on the CHROMOMETHYLASE3 gene from Arabidopsis thaliana. The sequence and variants come from GenBank, gi|14647156; the homology model derives from two blocks families, IPB001525:C5_DNA_meth and IPB002857:Znf-CXXC, found by a Reverse PSI-BLAST search of the Blocks database. In the ‘Genomic Sequence’ plot the top region of the graphics shows the locations of the blocks on the reference sequence; the first four correspond to blocks IPB001525 A through D in order; the final block corresponds to IPB002857 F. The middle shows the locations of the exons, represented by boxes. The bottom region displays the locations of the polymorphisms; the first row of triangles displays truncation changes in red, as they are most likely to be deleterious, the second row displays missense changes in black and the third row (not present in this figure) would show silent changes in purple. Upward-pointing triangles indicate an exonic change, while downward-pointing triangles indicate an intronic change. The ‘Coding Sequence’ plot positions the same information on the spliced coding sequence.
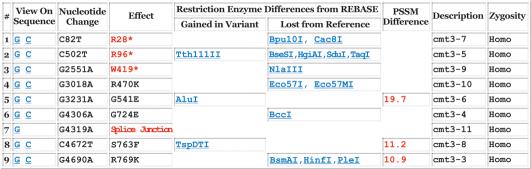
Figure 2.
Table of polymorphisms for the gene shown in Figure 1. The ‘View on Sequence’ column provides links to the detailed sequence display, shown in Figure 3; G links to the genomic sequence and C links to the coding sequence. The restriction enzyme names link to entries in REBASE, describing commercial availability and isoschizomers. Descriptions are extracted from the GenBank entry. The last column shows the zygosity of the change; if a variant had been entered using an ambiguous nucleotide, this column would read ‘hetero.’
Figure 3.
Polymorphisms on the sequence from Figures 1 and 2. This region shows a block aligned on the reference sequence (the underlined regions), two missense changes (numbers 6 and 8 in Fig. 2), a splice junction change in red (number 7) and two introns (the regions which are not spaced in triplets). The second missense change is colored red because the ΔPSSM score is >10. The residues of the reference protein are colored to indicate how each position compares to the aligned block; those residues colored green are most similar to the corresponding column in the block, while those colored red are most diverged.
AVAILABILITY
PARSESNP can be accessed over the web at http://www.proweb.org/parsesnp/. A user's guide is provided at http://www.proweb.org/parsesnp/parsesnp_help.html. In addition, the source code to PARSESNP is available as part of a suite of programs from http://www.proweb.org/.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Pauline Ng for providing a program to calculate the information content of blocks, Jorja Henikoff for help developing the software, Steve Henikoff for helpful suggestions which have improved the user interface and Jorja and Steve for their critical reading of the manuscript. This work was funded by the Department of Energy Office of Biological and Environmental Research and the National Science Foundation Plant Genome Research Program. N.E.T. was supported in part by the Howard Hughes Medical Institute.
REFERENCES
- 1.Krawczak M. and Cooper,D.N. (1997) The human gene mutation database. Trends Genet., 13, 121–122. [DOI] [PubMed] [Google Scholar]
- 2.Wheeler D.L., Church,D.M., Federhen,S., Lash,A.E., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Sequeira,E., Tatusova,T.A. and Wagner,L. (2003) Database resources of the National Center for Biotechnology. Nucleic Acids Res., 31, 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colbert T., Till,B.J., Tompa,R., Reynolds,S., Steine,M.N., Yeung,A.T., McCallum,C.M., Comai,L. and Henikoff,S. (2001) High-throughput screening for induced point mutations. Plant Physiol., 126, 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ulrich C.M., Bigler,J., Sibert,J., Greene,E.A., Sparks,R., Carlson,C.S. and Potter,J.D. (2002) Cyclooxygenase 1 (COX1) polymorphisms in African-American and Caucasian populations. Hum. Mutat., 20, 409–410. [DOI] [PubMed] [Google Scholar]
- 5.Till B.J., Reynolds,S., Greene,E.A., Codomo,C., Enns,L., Johnson,J., Burtner,C., Odden,A., Young,K., Taylor,N. et al. (2003) Large-scale discovery of induced point mutations with high throughput TILLING. Genome Res., 13, 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M.C., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O'Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts R.J. and Macelis,D. (2001) REBASE—restriction enzymes and methylases. Nucleic Acids Res., 29, 268–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henikoff J.G., Greene,E.A., Pietrokovski,S. and Henikoff,S. (2000) Increased coverage of protein families with the blocks database servers. Nucleic Acids Res., 28, 228–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henikoff S., Henikoff,J.G., Alford,W.J. and Pietrovski,S. (1995) Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene, 163, 17–26. [DOI] [PubMed] [Google Scholar]
- 11.Henikoff S. and Henikoff,J.G. (1994) Protein family classification based on searching a database of blocks. Genomics, 19, 97–107. [DOI] [PubMed] [Google Scholar]
- 12.Henikoff J.G. and Henikoff,S. (1996) Using substitution probabilities to improve position-specific scoring matrice. Comput. Appl. Biosci., 12, 135–143. [DOI] [PubMed] [Google Scholar]
- 13.Bailey T.L. and Gribskov,M. (1998) Combining evidence using p-values: application to sequence homology searches. Bionformatics, 14, 48–54. [DOI] [PubMed] [Google Scholar]
- 14.Ng P.C. and Henikoff,S. (2001) Predicting deleterious amino acid substitutions. Genome Res., 11, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider T.D., Stormo,G.D., Gold,L. and Ehrenfeucht,A. (1986) Information content of binding sites on nucleotide sequences. J. Mol. Biol., 188, 415–431. [DOI] [PubMed] [Google Scholar]
- 16.Altschul S.F. (1991) Amino acid substitution matrices from an information theoretic perspective. J. Mol. Biol., 219, 555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]