Abstract
We present four tools for the analysis of RNA secondary structure. They provide animated visualization of multiple structures, prediction of potential conformational switching, structure comparison (including local structure alignment) and prediction of structures potentially containing a certain kind of pseudoknots. All are available via the Bielefeld University Bioinformatics Server (http://bibiserv.techfak.uni-bielefeld.de).
INTRODUCTION
The Bielefeld University Bioinformatics Server (BiBiServ) supports internet-based collaborative research and education in bioinformatics. It was established in 1996 and currently provides a variety of 15 software tools (summarized in Table 1), together with a repository of educational material. These resources are accessible at http://bibiserv.techfak.uni-bielefeld.de. This article restricts itself to four tools offered for the analysis of RNA secondary structure.
Table 1. BiBiServ tool synopsis.
| Name | Purpose | Availability | Input | Output |
|---|---|---|---|---|
| Genome comparison | ||||
| AGenDa | Align-based gene detection | B | DNA sequence | Exon list, alignment graphics |
| MGA | Multiple Genome Aligner | D | DNA sequence | Alignment |
| REPuter | Fast computation of repeats in complete genomes | B, D | DNA sequence | Repeats and their position |
| Alignments | ||||
| AltAVisT | Alternative Alignment Visualization Tool | B | DNA-/protein sequence | Alignment |
| DCA | Divide-and-Conqueror Multiple Alignment | B, D | DNA-/protein sequence | Alignment |
| DIALIGN | Multiple sequence alignments | B, D, E | DNA-/protein sequence | Alignment |
| JALI | Jumping Alignments | B,D | Protein sequences and multiple alignment | Alignment |
| TSDA | Pairwise alignment of recombinant DNA sequences | B,D | Scoring parameter, DNA sequences | Alignment |
| Primer design | ||||
| GeneFisher | Design of degenerate PCR primers; also for single primer PCR | B | Unaligned DNA-/protein sequence | Primer sequences |
| RNA-studio | ||||
| paRNAss | Prediction of Alternate RNA Secondary Structures | B | RNA sequence | Predicted confirmations |
| pknotsRG | RNA folding and thermodynamic matching; including pseudoknots | B | RNA sequence | MFE structure, best pseudoknot |
| RNA Forester | RNA structure alignment and motif discovery | B | RNA secondary structures | Global/local structure alignment |
| RNA Movies | Animated visualization of a series of RNA secondary structures | D | Movie script | Structure graphics, animation |
| Evolutionary relationship | ||||
| ROSE | Random-model Of Sequence Evolution | B, D | Model parameter, required data size | Sequence families |
| SplitsTree | Distance-based phylogeny reconstruction based on split decomposition | B, D, E | Matrix of pairwise distances | Split decomposition graph |
Availability code: B, available as BiBiServ WWW Tool; D, available for download; E, also available elsewhere.
THE BiBiServ RNA STUDIO
The BiBiServ RNA Studio is a collection of tools for the analysis of RNA secondary structure. Most of the tools are dedicated to dealing with more than just one structure in one way or another. They expect structures determined experimentally, delivered by comparative studies or predicted by other programs like MFOLD (1) or RNAfold (2). While it is planned to integrate these tools more closely in the future, currently it is the user's responsibility to channel the data through a multi-step analysis.
There are many occasions where one has to deal with a set of related RNA secondary structures: analyzing a set of near-optimal structures, the folding path of an RNA molecule or the reshaping of a structure that acts as a conformational switch. Comparing structures by merely looking at them is rather difficult. RNA Movies is a system for the visualization of multiple RNA secondary structures in the form of animated structure graphics. In an animated series of structures, common features stand still while the eye is caught by those who change. RNA Movies thus creates the impression of an RNA-molecule moving through its own 2D structure space. This dynamic effect can only be presented imperfectly on paper (Fig. S1 in the Supplementary Material shows an animation of a potential re-folding path of a conformational switch in the ribosomal protein S15 mRNA of Escherichia coli).
The visualization is presented in the paradigm of a video-player, providing buttons for moving forward, backward, single step etc., combined with layout options (labelled/unlabelled, bonds, computer screen/public presentation, static screenshot as GIF or postscript). The speed of the animation is controlled by adjusting the number of interpolations computed between two successive structures.
Doing pure visu1alization, RNA Movies is not responsible for the content it shows. Movie scripts are generated by other programs or can even be written by hand. The format uses plain text and structures are written in the dot-parenthesis notation also used in the RNAfold package. There are two versions of RNA Movies: the C version uses the Mesa graphics library and is available for download only. The WWW version, written in Java, is currently under revision.
There are various cases where the biological function of an RNA molecule involves a reversible change of conformation. paRNAss is a software approach to predict the potential of conformational switching in RNA. It is based on three hypotheses about the secondary structure space of a switching RNA molecule, which can be evaluated by RNA folding and structure comparison. In the positive case, the predicted structures must be verified experimentally.
In order to be predicted as a potential conformational switch, the folding space of an RNA molecule must satisfy the following conditions:
There must be two local minima in the folding space close to the energetic optimum. They must be significantly different, as they represent two positions of the switch with different regulatory function.
The structures residing in these minima must be clearly separated by an energy barrier, to ensure that each confirmation is stable and switching is triggered by external events.
The folding space must not provide another (third) local minimum, as we assume that the RNA automatically finds the alternative position once the change is triggered.
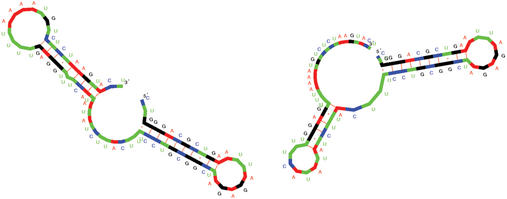
Note that the folding space can contain tens or hundreds of local minima. Only the last condition brings about statistical significance and it is the essential feature checked by paRNAss. This is done by sampling the near-optimal solution space, and evaluating the pairwise similarities of the structures. This sampling is expensive, but necessary, since RNA folding programs cannot compute local minima of the folding space only. Three different notions of structural similarity are used. Figure 1 (and Fig. S2 in the Supplementary Material) shows the two structures predicted for S15 mRNA of E.coli, known to be a conformational switch (5). Figure S1 in the Supplementary Material visualizes a possible transition path between the two alternative conformations as movie.
Figure 1.
Two structures predicted by paRNAss for S15 mRNA of E.coli (X01073), known to be a conformational switch (5).
While there are several programs available that compare two RNA secondary structures and compute a distance or a similarity score, none of them produces a structure alignment. This is the purpose of RNA Forester. RNA structures are represented as trees or more generally, sequences of trees, called forests. The alignment model underlying RNA Forester is the mathematically faithful generalization of sequence alignments to trees.
Global and local similarity alignments of RNA structures can be computed by a new algorithm that generalizes the Smith–Waterman algorithm from sequence to tree data. It finds the most similar substructure within two arbitrary structures. Hence it can discover conserved structural motifs in the absence of sequence conservation. Note that the best conserved substructures need not be aligned to each other in the best global alignment of two structures, hence the local alignment option is the more versatile technique.
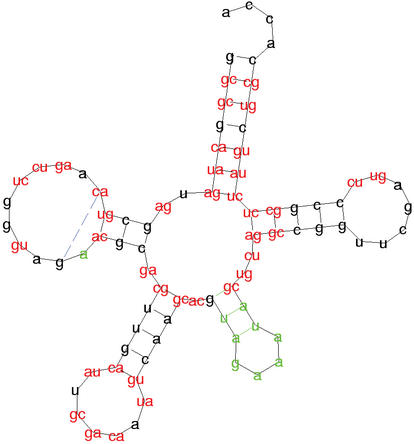
Dynamic programming over a tree domain is more sophisticated than over sequences, and the implementation uses carefully designed indexing schemes to achieve good space and time efficiency. The resulting alignments are drawn in a color code, highlighting common features, deviations, and compensatory base changes. Figure 2 shows a global alignment of the E.coli alanin and leucin tRNA structure as computed by RNA Forester.
Figure 2.
RNA Forester: squiggle-plot of the structure alignment of the tRNAs for alanine and leucine. Bases printed in black show structure elements that occur in both structures with the same sequence. Sequence variations are displayed by using red letters. Bases or base pairs that can only be found in alanine are printed in blue, while bases that only occur in leucine are printed in green.
Computing the RNA structure of minimal free energy (MFE) including pseudoknots in the standard thermodynamic model has been shown to be an NP-complete problem. Hence, pseudoknots will remain excluded from standard RNA folding programs, in spite of the fact that they are structural features of great interest. The specialized program pknotsRE by Rivas and Eddy (6) requires O(n6) time and O(n4) space to recognize the class of chained simple recursive pseudoknots. The BiBiServ tool pknotsRG restricts further to the class of canonical simple recursive pseudoknots, still general enough to cover many cases of pseudoknots reported in the literature. It achieves efficiency O(n4) time and O(n2) space, allowing to fold sequences up to 400 nt within 1–2 h.
The longer an RNA molecule, the more important the actual folding path and the less we can rely on the MFE structure predicted by folding programs. The MFE structure may not contain the pseudoknot actually present in the native structure. The program provides the option to compute the best pseudoknot contained in any feasible structure for the input RNA sequence, where ‘best’ is defined as minimal value of free energy for a pseudoknot substructure, divided by the length of sequence involved in the pseudoknot.
Figure 3 shows the text output returned by pknotsRG for the pseudoknot in the RNA sequence of turnip yellow mosaic virus. A pseudoknot is indicated by square and curly brackets.
Figure 3.
Text output from pknotsRG for the pseudoknot in the RNA sequence of turnip yellow mosaic virus.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
| RNA Movies | Animated visualization of a series of RNA secondary structures |
| Input: | A movie ‘script’ in text format |
| Output: | Structure graphics, animation, screenshots as GIFs or postscript |
| Performance: | O(kn), no apparent limits |
| Tool authors: | Dirk Evers |
| References: | (3) |
| URL | http://bibiserv.techfak.uni-bielefeld.de/rnamovies/ |
| paRNAss | Prediction of Alternating RNA Secondary Structures |
| Input: | RNA sequence |
| Output: | Hypothetical switch conformations, transition path as RNA Movie |
| Performance: | O(n3+n2s2), where s is the sample size |
| 75 s for a sequence of length 75 and sample size 50 | |
| Tool authors: | Dirk Haase, Marc Rehmsmeier, Robert Giegerich |
| References: | (4) |
| URL | http://bibiserv.techfak.uni-bielefeld.de/parnass/ |
| RNA Forester | RNA structure alignment and motif discovery |
| Input: | Two or more RNA secondary structures (Vienna notation) |
| Output: | Global (pairwise or multiple) or local structure alignment and similarity score/distance |
| Performance: | O(|F1||F2|deg(F1)deg(F2)(deg(F1)+deg(F2))), where |Fi| is the number of nodes in forest Fi and deg(Fi) is the degree of Fi, for i=1,2. |
| 10 s for two sequences of length 235 | |
| 3.35 min for two sequences of length 550 | |
| Tool authors: | Matthias Höchsmann |
| References: | In preparation |
| URL | http://bibiserv.techfak.uni-bielefeld.de/rna-forester/ |
| pknotsRG | RNA folding and thermodynamic matching, including pseudoknots |
| Input: | RNA sequence |
| Output: | MFE-structure (knotted or not), or best pseudoknot |
| Performance: | O(n4) time, O(n2) space |
| 15 s/9 Mb for sequence of length 100 | |
| 1.20 h/256 Mb for sequence of length 400 | |
| Tool authors: | Jens Reeder, Robert Giegerich |
| References: | In preparation |
| URL | http://bibiserv.techfak.uni-bielefeld.de/pknotsrg/ |
Acknowledgments
ACKNOWLEDGEMENTS
We acknowledge the continuing support of our networking group, Peter Koch and Rainer Orth, and the work of Kai Runte and Michael Höhl who have served several years as BiBiServ administrators. Thanks to Ute Willhoeft and Burkhard Morgenstern for valuable suggestions to improve previous versions of this manuscript.
REFERENCES
- 1.Zuker M., Mathews,D.H. and Turner,D.H. (1999) Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In Barciszewski,J. and Clark,B.F.C. (eds), RNA Biochemistry and Biotechnology, NATO ASI Series, Kluwer Academic Publishers, pp. 11–43. [Google Scholar]
- 2.Hofacker I.L., Fontana,W., Stadler,P.F., Bonhoeffer,L.S., Tacker,M. and Schuster,P. (1994) Fast folding and comparison of RNA secondary structures. Monatsh. Chem., 125, 167–188. [Google Scholar]
- 3.Evers D. and Giegerich,R. (1999) RNA movies: visualizing RNA secondary structure spaces. Bioinformatics, 15, 32–37. [DOI] [PubMed] [Google Scholar]
- 4.Giegerich R., Haase,D. and Rehmsmeier,M. (1999) Prediction and visualization of structural switches in RNA. In Proc. 1999 Pacific Symposium on Biocomputing, World Scientific, pp. 126–137. [DOI] [PubMed] [Google Scholar]
- 5.Philippe C., Benard,L., Portier,C., Westhof,E., Ehresmann,B. and Ehresmann,C. (1995) Molecular dissection of the pseudoknot governing the translational regulation of Escherichia coli ribosomal protein S15. Nucleic Acids Res., 23, 18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rivas E. and Eddy,S.R. (1999) A dynamic programming algorithm for RNA structure prediction including pseudoknots. J. Mol. Biol., 285, 2053–2068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.