Abstract
Several primer prediction and analysis programs have been developed for diverse applications. However, none of these existing programs can be directly used for the design of primers in protein interaction experiments, since proteins may have transmembrane domains (TMDs) and/or a signal peptide that must be excluded from experiments. Furthermore, it is frequently the case that a short restriction sequences must be added to each primer in order to clone PCR products into a given destination vectors for expression. DePIE, a web-based primer design tool, was developed to address these deficiencies. The program takes as input NCBI protein accession numbers and returns primer information including nucleotide sequences, thermodynamic melting temperature of the nucleotide sequences and the target positions. DePIE is implemented in JAVA, PERL and PHP and has proven to be very efficient in designing primers for our interaction experiments. DePIE services can be accessed at the web site: http://biocore.unl.edu/primer/primerPI.html.
INTRODUCTION
The design of PCR and sequencing primers is an essential activity of molecular biology. A variety of primer prediction and analysis programs have been developed to determine primers for diverse applications (1). Many of the programs take as input a set of sequences and select candidate single primers or primer pairs based on melting temperature properties and secondary structure behavior including hairpin formation, self-hybridization and cross-dimerization. This ensures the availability of the primer for the reaction as well as minimizing the formation of primer dimers.
Recently, research has increasingly focused on the use of yeast two-hybrid assays to screen in a high-throughput fashion for protein–protein interactions (2,3). Identifying and ultimately understanding these protein–protein interactions is a critical step in functional genomic analysis (4). A pivotal step to the success of the two-hybrid assay is the design of suitable primers. Existing primer prediction programs however cannot be directly used to design primers in this context for the following reasons. Firstly, proteins may have a signal peptide and/or transmembrane domains (TMDs) that must be excluded from the experiments, since these regions do not participate in the interaction between proteins. Secondly, it is usually necessary to add a short restriction or recombination sequence to each primer in order to facilitate the cloning of PCR products into yeast two-hybrid bait and prey vectors for expression. Thirdly, existing programs do not easily lend themselves to a high-throughput approach for yeast two-hybrid assay projects. In particular, there are several steps that need to be automated: (i) both the nucleic and amino acid sequences must be retrieved from relevant databases on-line; (ii) several TMD and structural prediction programs must be applied to each such sequence; and (iii) start/stop sites in the nucleic acid sequences must be determined before applying the actual primer design package.
To address each of these shortcomings, we have created DePIE, a web-based primer design program used for protein interaction experiments. We describe the basic architecture of the system below.
DESIGN AND IMPLEMENTATION
DePIE is a web tool designed using UML (Unified Modeling Language) in Rational Suite® DevelopmentStudio® (http://www.rational.com/products/dstudio/index.jsp) and implemented in the Java programming language in SunTM ONE Studio 4 (http://wwws.sun.com/software/product_categories/development_tools.html). Both PHP and Perl are used by the web interface and query-parsing scripts.
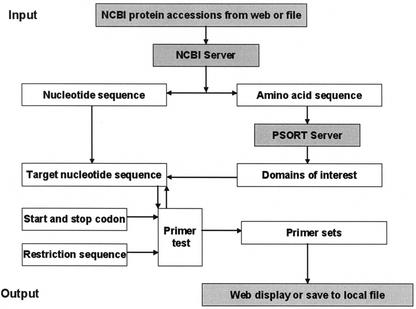
The pipeline of data processing is demonstrated in Figure 1 and detailed as follows. Firstly, both the DNA and amino acid sequences are retrieved from GenBank using Entrez, a sequence retrieval system developed at NCBI (http://www.ncbi.nlm.nih.gov/). The amino acid sequence is then used to predict the structure and topology of the corresponding proteins with the PSORT program (5). The resulting HTML page generated by the PSORT server is then parsed to get information about the signal peptide, transmembrane domains and topology. Based on the information from the PSORT, the domains of interest (for example, domains within the endoplasmic reticulum), are determined and their corresponding start and end positions are calculated. For each domain of interest, an 18-base nucleotide sequence is retrieved from each end of its corresponding nucleotide sequence. The start and end codons are added a priori to the 5′ end of the forward and reverse 18-base primers, respectively. Thus, each primer is composed of 21 bases. Protein interaction experiments usually require the cloning of PCR products into a given vectors for expression. It is thus necessary to have an option for the user to input short sequences with restriction or recombination sites that will be added priori to the 5′ end of primer segments. Since GATEWAYTM provides an extremely fast and efficient route for functional analysis of genes, protein expression and cloning or subcloning of DNA fragments (6,7), we set the restriction sequences used to build GATEWAYTM clones by default. Their sequences are GGGGACAAGT-TTGTACAAAAAAGCAGGCTCT for the forward primer and GGGGACCACTTTGTACAAGAAAGCTGGGTN for the reverse primer, respectively. These default values can be changed when required. Other short sequences can be added in order to design primers for direct entry of the PCR product into other vector series (i.e. other yeast two-hybrid vector systems or other fusion protein expression systems).
Figure 1.
The pipeline of data processing in DePIE. Refer to the text for a detailed description.
The candidate primers must meet the following requirements: (i) primer sequences should have 35–65% G-C content; (ii) the annealing temperature of each primer should match and be within a 45–75°C range; (iii) the primer should be able to form ‘G/C’ clamps. The bonds between G and C will facilitate the initiation of complementary strand formation by Taq polymerase acting at the 3′ end of the hybridized primer (8); (iv) at the 3′ end, there are not three or more G or C bases; (v) primers should not have a high tendency to form secondary structure; (vi) mispriming, i.e. hybridization of one or more non-target regions, should be avoided. Reasons why the above-mentioned rules need to be considered have been described in Rozen and Skaletsky (1) and Lowe et al. (8). DePIE uses an annealing test described in Hillier and Green (9), with slight modification, to check individual primers for self-complementarity and to check the two primers in a PCR primer pair for complementarity to each other.
If all of the above conditions are satisfied by a candidate pair, the program outputs primer information and other options. Otherwise the program replaces the unsatisfied segment with another 18-base segment, which moves the window inward three bases (i.e. there is only 3-base difference between the new segment and the replaced one). This operation is repeated until the candidate pair satisfies all of the above requirements or the sequence is exhausted.
INPUT, OUTPUT AND PERFORMANCE
The input for DePIE consists of NCBI protein accession numbers. The accession numbers can be entered manually in the web page or uploaded by file from a local computer. The output for DePIE includes the primer sequences, TM (thermodynamic melting point temperature), and target positions within the nucleotide sequence. Other options include the nucleotide sequences, amino acid sequences and TMP/protein topology predication results. The output is returned as a HTML file and a text file can be saved locally.
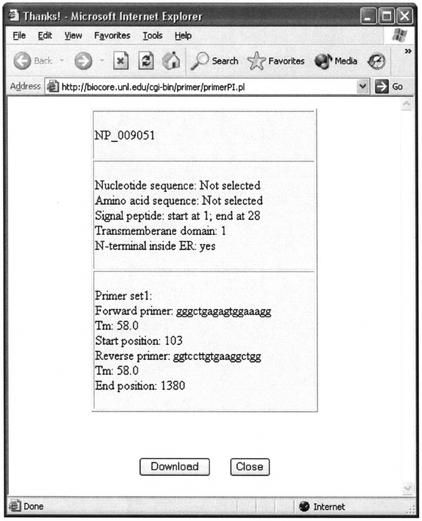
Figure 2 shows a sample output produced by DePIE. Note that the output option, PSORT predictions, was selected. The protein with NCBI accession number NP_009051 is predicted to have a signal peptide (the first 28 amino acids), a transmembrane domain and N-terminal inside endoplasmic reticulum. Primer sequences are gggctgagagtggaaagg for the forward and ggtccttgtgaaggctgg for the reverse, respectively. Thermodynamic melting point temperature is 58°C for both primers. The targeting PCR product is from base 103 to 1380 within the nucleotide sequence.
Figure 2.
RNA Forester: squiggle-plot of the structure alignment of the tRNAs for alanine and leucine. Bases printed in black show structure elements that occur in both structures with the same sequence. Sequence variations are displayed by using red letters. Bases or base pairs that can only be found in alanine are printed in blue, while bases that only occur in leucine are printed in green.
DePIE is currently being used in our laboratory and has so-far proven to be very efficient in designing primers for our interaction experiments. The testing of results and experimental findings will be reported in a separate paper.
DISCUSSION AND CONCLUSION
We implemented a web-based primer design program for protein interaction experiments, which we refer to as DePIE. Users can use this tool via any common web browser using a variety of operating systems. The salient feature of DePIE is that it makes use of existing web resources, e.g. the NCBI database and the PSORT protein prediction tool, and implemented an automated pipeline of designing primers for protein interaction experiments. Another important feature which makes DePIE different from other primer design programs is that it provides an option to add a short restriction sequences prior to the primers. It is essential in the expression experiment step for PCR products to be cloned into destination vectors for expression. Although it takes as default the sequences used to build GATEWAYTM clones, DePIE could broaden its application to other protein expression systems that would involve amplifying full length genes.
The bioinformatics tool reported in this paper would benefit future proteomic research. As shown in Figure 1, DePIE makes use of existing bioinformatics tools to predict domains of interest. We can take the advantage of diverse protein interaction databases and evolutionary conserved domain databases to create domain interaction profiles (10). Based on this, we can apply computer algorithms, for example, Support Vector Machine (SVM) and Hidden Markov Model (HMM), to infer interactions of the domains which are found by DePIE (11,12). Putting together the knowledge learned from protein domain binary interactions predicted, we might be able to build up a network of protein–protein interactions in either an organelle, e.g. endoplasmic reticulum or an organism, e.g. Saccharomyces cerevisiae, of interest (13,14).
DePIE accepts as input only NCBI protein accession numbers in the current version. It is expected to extend the capabilities of DePIE to be able to accept accession numbers of other biological databases, e.g. EMBL and SWISS-PROT, nucleotide sequences and peptide sequences. In addition, since DePIE needs to communicate with other web servers, i.e. NCBI server and the PSORT server, the most expensive operation is data transportation between web servers. Its running time may be a concern for web interface users. This is acutely problematic if such servers are not functioning. Therefore, we plan to releases a version of our software in the near future that allows PSORT and the NCBI database to be accessed locally.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Bin Ge, Kaleigh Smith and Michelle Scott for helpful discussions and suggestions. We thank The Nyguan, Yavuz Yavuz and Qiaomei Zhong for their help in making DePIE accessible on the web. The support from Genome Quebec (http://www.genomequebec.com/) is gratefully acknowledged.
REFERENCES
- 1.Rozen S. and Skaletsky,H.J. (2000) Primer3 on the WWW for general users and for biologist programmers. In Krawetz, S. and Misener, S. (eds), Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, NJ, pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 2.Ito T., Tashiro,K., Muta,S., Ozawa,R., Chiba,T., Nishizawa,M., Yamamoto,K., Kuhara,S. and Sakaki,Y. (2002) Toward a protein–protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc. Natl Acad. Sci USA, 97, 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uetz P., Giot,L., Cagney,G., Mansfield,T.A., Judson,R.S., Knight,J.R., Lockshon,D., Narayan,V., Srinivasan,M., Pochart,P. et al. (2000) A Comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]
- 4.Eisenberg D., Marcotte,E.M., Xenarios,I. and Yeates,T.O. (2000) Protein function in the post-genomic era. Nature, 405, 823–826. [DOI] [PubMed] [Google Scholar]
- 5.Nakai K. and Horton,P. (1999) PSORT: a program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem. Sci., 24, 34–35. [DOI] [PubMed] [Google Scholar]
- 6.Hartley J., Temple,G. and Brasch,M.A. (2000) DNA cloning using in vitro site-specific recombination. Genome Res., 10, 1788–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walhout A.J.M., Temple,G.F., Brasch,M.A., Hartley,J.L., Lorson,M.A., van den Heuvel,S. and Vidal,M. (2000) GatewayTM recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol., 328, 575–592. [DOI] [PubMed] [Google Scholar]
- 8.Lowe T.M., Sharefkin,J., Yang,S.Q. and Dieffenbach,C.W. (1990) A computer program for selection of oligonucleotide primers for the polymerase chain reaction. Nucleic Acids Res., 18, 1757–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hillier L. and Green,P. (1991) OSP: an oligonucleotide selection program. PCR Methods Applic., 1, 124–128. [DOI] [PubMed] [Google Scholar]
- 10.Xenarios I. and Eisenberg,D. (2001) Protein interaction databases. Curr. Opin. Biotechnol., 12, 334–339. [DOI] [PubMed] [Google Scholar]
- 11.Bock J.R. and Gough,D.A. (2001) Predicting protein–protein interactions from primary structure. Bioinformatics, 17, 455–460. [DOI] [PubMed] [Google Scholar]
- 12.Deng M., Mehta,S., Sun,F. and Chen,T. (2002) Inferring domain–domain interactions from protein–protein interactions. Genome Res., 12, 1540–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong A.H., Drees,B., Nardelli,G., Bader,G.D., Brannetti,B., Castagnoli,L., Evangelista,M., Ferracuti,S., Nelson,B., Paoluzi,S. et al. (2002) A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science, 295, 321–324. [DOI] [PubMed] [Google Scholar]
- 14.Schwikowski B., Uetz,P. and Fields,S. (2000) A network of protein–protein interactions in yeast. Nat. Biotechnol., 18, 1257–1261. [DOI] [PubMed] [Google Scholar]