Abstract
STING Millennium Suite (SMS) is a new web-based suite of programs and databases providing visualization and a complex analysis of molecular sequence and structure for the data deposited at the Protein Data Bank (PDB). SMS operates with a collection of both publicly available data (PDB, HSSP, Prosite) and its own data (contacts, interface contacts, surface accessibility). Biologists find SMS useful because it provides a variety of algorithms and validated data, wrapped-up in a user friendly web interface. Using SMS it is now possible to analyze sequence to structure relationships, the quality of the structure, nature and volume of atomic contacts of intra and inter chain type, relative conservation of amino acids at the specific sequence position based on multiple sequence alignment, indications of folding essential residue (FER) based on the relationship of the residue conservation to the intra-chain contacts and Cα–Cα and Cβ–Cβ distance geometry. Specific emphasis in SMS is given to interface forming residues (IFR)—amino acids that define the interactive portion of the protein surfaces. SMS may simultaneously display and analyze previously superimposed structures. PDB updates trigger SMS updates in a synchronized fashion. SMS is freely accessible for public data at http://www.cbi.cnptia.embrapa.br, http://mirrors.rcsb.org/SMS and http://trantor.bioc.columbia.edu/SMS.
INTRODUCTION
SMS was designed in order to meet exciting new challenges originated specifically from the recent rise in biologists' interest for describing and quantifying the relationship between sequence, structure and the function of proteins. This sudden rise in interest occurs in parallel to a disproportional growth between sequence and structure databases. Knowing that well organized information about macromolecular sequence, structure and function is an important platform for understanding processes in living organisms and diseases, as well as for designing new drugs, biologists search for appropriate tools to crosslink and analyze (often heterogeneous) data that have already been collected. Such tools should help elucidating patterns and underlying associations hidden in original raw data. To date, the structure of over 20 000 macromolecules and their complexes has been determined and deposited at the Protein Data Bank (PDB) (1). By calculating sequence–structure–function descriptors from experimental databases we may establish a subtle link between key sequence patterns and structure/function attributes. In addition, by identifying similarities in folds from alignments of protein chains we might be able to find a common base that defines function.
Protein function determination and identification of possible function modifiers, is in fact the most desirable goal of many genome project efforts. To achieve such goal, bioinformatics counts with some basic activities, such as:
sequence homology search and position specific sequence conservation;
sequence alignments;
structural alignments;
search for sequence to structure relationship;
definition of structural parameters like intra and inter chain contacts;
structure modeling with evaluation of the quality of obtained models;
protein interfaces and identification of active and ligand binding sites;
cumulative statistics on protein family characteristics.
All these activities require an integrated environment with computational tools that can visually and schematically map 3D information together with parameters and the patterns extracted from a number of databases, on the protein sequence. In some other software packages, for multi-parameter presentation, the key role is trusted to simple structure display. Although the molecular structure display and manipulation is becoming an everyday task not only to structural biologists, but to an increasing audience of molecular biologists and biochemists as well, multi-parameter mapping on the structure itself is intrinsically limited. This is because any point on the structure/surface might be painted (colored) only once, following the color code designed for given amino acid characteristics. Amino acid sequence on the other hand, can be mapped by number of parameters, organized and displayed in underlying rows. Appropriate visualization of numerical value for any given parameter and for any amino acid in the sequence, might be easily achieved by color coding. This is the crucial difference between existing software packages (2,3) on one side and our SMS component—Protein Dossier.
The constant increase in accumulation of sequence data prompts for a more complete functional characterization of the gene product. This necessarily leads to insights into the protein's three-dimensional (3D) structure. Given this scenario, it is of utmost importance that the programs designed to provide most complete and versatile information through user friendly interface, are as intuitive as possible. Such programs ought to save the end-user from dealing with a complicated sequence of programming commands typically required in order to achieve complex data cross-referencing. This task is only possible at the expense of an elaborate user interface. STING Millennium Suite is the mature product, the results of our attempts to meet such challenging requirements.
STING Millennium Suite INTRINSICS
STING is an acronym of Sequence To and withIN Graphics. The original STING program was developed to enable bi-directional coupling between sequence and structural information and also to offer a simple and easy way to map a single amino acid (or nucleotide) letter code to its position in 3D, and vice versa. This work was originally published elsewhere (4).
STING Millennium was developed from the original STING concept. However, we have completely rewritten all code and made the STING concept much more versatile.
SMS version 2.2 is now available from the network of mirrors: http://www.cbi.cnptia.embrapa.br, at EMBRAPA/CNPTIA, Campinas-SP, Brazil, http://mirrors.rcsb.org/SMS, at PDB and http://trantor.bioc.columbia.edu/SMS, at Columbia University in Barry Honig's laboratory. Other SMS mirror sites are anticipated and some are in the final phase of implementation. SMS entry page clearly indicates current SMS version, SMS host server (city and country) and three access options: (i) SMS web page illustrated with art images and which is best seen if using a fast internet connection and high resolution monitors; (ii) SMS access web page similar to the one described above, but with alternative molecular image illustrations; and (iii) SMS simple web page designed for access by users having low resolution monitors and slow internet connections. SMS web page menu options have been designed and written in such a way so that a user may quickly find critical information about each SMS component and possible interpretation of presented data.
In terms of the system design the package is implemented using a variety of programming languages, each one better suited to a specific task: (i) the JavaTM language is used for the SMS Sequence window, which allows a complete control of the Chime plugin as well as initiation of any of the SMS components; (ii) the JavaScript is used to make the communication between Java Sequence window and Chime-plugin; (iii) the Perl language is used as a wrapper for processing web requests through CGI; (iv) the C++ language is used for more intensive programming tasks in structure parameter calculations; and (v) Chime® molecular visualization plugin (MDL inc.) is used to provide the molecular rendering.
SMS ORGANIZATION
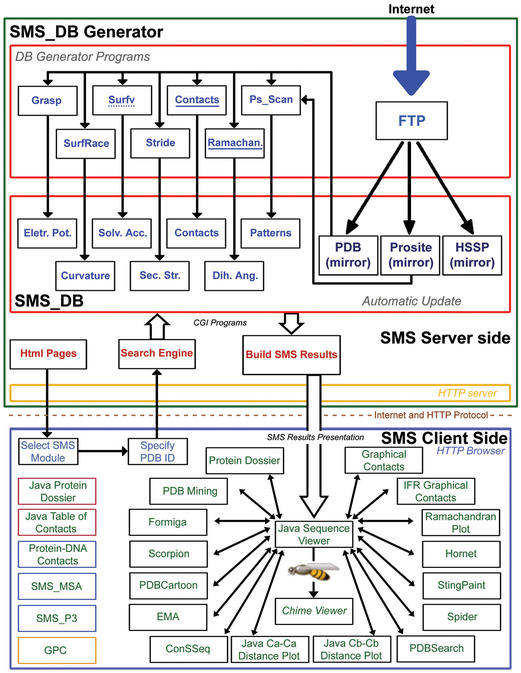
SMS is organized in two logical layers (Fig. 1): SMS server and SMS client. The server side is responsible for updating regularly all relevant public domain databases used by SMS: PDB, HSSP (5–7) and PROSITE (8). At the same time, the SMS server is also responsible for calculation of a number of macromolecular properties for each PDB structure: electrostatic potential is calculated using GRASP (9) software, curvature is calculated using SurfRace (10) software, solvent accessible area for each protein chain and for the whole molecular complex is calculated using Surfv (11) software adapted to our own requirements, secondary structure identification is calculated according to DSSP (12) and STRIDE (13), intra and inter chain amino acid contacts as well as protein/dna interaction are calculated by our own software: contacts, hydrophobicity is assigned according to Radzicka and Wolfenden (14), dihedral angles are calculated by our own ‘Ramachan’ program and PROSITE patterns are identified using Ps_Scan (15) software. SMS components written in italic are accessible in SMS 3.0, now available on our beta test site, and SMS components written in italic and underlined are to be available in SMS 3.1 version. Server side is also responsible for providing access to SMS database through HTTP protocol (16). The SMS client side provides a user friendly graphical interface and communicates to SMS server, sending user's requests and receiving SMS responses. The interaction between SMS client and server happens in two steps. (i) A HTTP client (web browser) allows a user to select any SMS module and also to specify a PDB ID. On the server side, a HTTP server listens to requests from SMS clients transmitted through the internet. When it receives a request from a client, it routes it to an appropriate CGI program—Common Gateway Interface. The CGI program searches for the selected PDB file and by accessing the SMS database, builds the appropriate SMS response to the request. (ii) A format of the SMS server response depends on which SMS module was selected by the user. For instance, if a user has selected Protein Dossier among the available SMS components, and then specifies PDB file name containing desirable protein structure, SMS search engine will collect corresponding data and a user will receive a cartoon containing several structural parameters mapped to each residue in the sequence. Resulting information presentation at the client side is done by using different resources such as HTML 3.2—Hyper Text Markup Language (17), javascript 1.2 (18) and java 2 (19) as well as the MDL Chime plugin (Chemscape Chime software, MDL information systems), available from Molecular Design Limited Inc. (MDL) at http://www.mdli.com/chemspace/chime.
Figure 1.
Schematic diagram illustrating the architecture of SMS. SMS is organized in two logical layers: SMS server and SMS client. On the server side, illustrated inside the large green box, are carried out all updates of the public domain databases used by SMS and the subsequent calculation of macromolecular properties. The SMS client side, illustrated inside the large blue box, provides a user friendly graphical interface and communicates to SMS server. The SMS versions 2.2, 3.0, 3.1 and 4.0 components are listed within black, red, blue and yellow border boxes, respectively.
In Figure 1, within the SMS client box, SMS version 2.2 components are listed within black border boxes, SMS version 3.0 components (now available at beta test site) are within red border boxes, SMS version 3.1 components (now at alpha test site) are within blue border boxes and finally SMS version 4.0 component (in final phase of development) is shown within yellow border box. SMS_MSA stands for the new SMS component: multiple sequence alignment. Those sequences are extracted both from HSSP and from our own SH2Qs (Sequences Homologue to the Query [Structure-having] Sequence). The latter one is designed so that we can benefit from the possibility of making a choice of the number of sequences that we want to consider in the relative entropy calculation. SMS_MSA will use standard MSA viewer package: SeaView (20). SMS_P3 sands for SMS phylogenetic tree component. Phylogenetic tree is generated starting from either HSSP or SH2Qs alignments. TreeView (21) is used to present to a user calculated phylogenetic trees. Finally, GPC is a new SMS component and it stands for global protein cartoon. This component displays, on a global sequence scale, a summary of a number of parameters which are presented in detail both in Protein Dossier and Java Protein Dossier.
STING Millennium MODES
STING Millennium has several working modes permitting its use with: (i) a single PDB structure (either from PDB or a single modeled structure in PDB format); (ii) a structural alignment of several protein structures [structures previously aligned with CE (22) or PRISM (23) software]; (iii) a PDB structure along with a sequence alignment, where the first sequence corresponds to the one with the known 3D structure. This SMS mode offers a way to visualize sequence conservation/variation and correlate it with structural regions in the target protein. The structure can also have its residues colored by an index that indicates sequence conservation (sequence relative entropy) promoting a rapid identification of the sites subject to differential (selective) evolutionary pressure.
STING Millennium MODULES
Several analytical programs developed by our group are listed as SMS components and are available under the modules menu in the SMS sequence frame (Fig. 2A). These SMS modules (components) perform a number of tasks providing a powerful combination of tools to analyze a given PDB structure. These modules (some are to be described in separate publications) are:
Ramachandran plot [SMS_RP] (24): a fully interactive java plot displaying the main-chain dihedral angles (φ and ψ). SMS_RP identifies the allowed/disallowed regions for the angles (Fig. 3F) and permits selection of residues that can be displayed in the SMS structure window.
PDB file TXT: lists the original PDB entry in text format.
Hornet: this SMS component does calculation and visualization of hydrogen bonds between two chains (protein and/or nucleic acid) with special attention given to water molecules (trapped between two chains) participating in H-bond formation.
Scorpion: this tool is designed to calculate the frequency of amino acid presence for a given (list of) PDB file(s) as well as frequency of amino acid neighbors in 3D.
Formiga: this SMS component calculates the frequency of amino acid neighbors in 3D, only from the interfacial areas between two or more protein chains, for a given (list of) PDB file(s) (Fig. 3G).
Graphical contacts and graphical IFR contacts: these two tools calculate atomic contacts between residues in a PDB entry. The complete profile of the interactions can be analyzed in the graphical contacts window and graphical IFR contacts window (Fig. 3D and E, respectively). Each of those windows displays the primary sequence of a chain and below it a histogram containing the number of contacts or IFR contacts made by the respective residue. Such contacts are divided into number of classes: hydrogen bonds (main-chain/main-chain, side-chain/main-chain, side-chain/side-chain), hydrogen bonds with intermediary water molecules, hydrophobic contacts, aromatic ring stacking contacts, electrostatic (attractive and repulsive) contacts and finally disulphide bridges. Each contact type is attributed with specific color both in histogram and in virtual contact lines presentation. Virtual contact lines connect residues of interest in a fan fashion. This presentation is very intuitive and the user can easily grasp contact parameters along the protein sequence. In the case of IFR contacts, IFRs are underlined with a red line for easy identification along the sequence. Both internal and IFR contacts are presented in tabular form as well (Fig. 3C).
StingPaint: visual aid to display multiple sequence alignments with the amino acids colored by physico-chemical properties. STINGpaint now supports the following sequence and Multiple Sequence Alignment (MSA) formats: (i) sequence of any PDB entry; (ii) sequence in FASTA format; (iii) MSA in CE and PRISM output format; (iv) MSA in ClustalW (25), PSI-BLAST (26) and GCG (27) output format.
PDB cartoon: image of the amino acid sequence along with the secondary structure elements rendered as cartoons.
PDB_Mining: this SMS component is designed to retrieve specific data from the PDB. This tool is an off-spin product, resulting from our compilation of scripts made to extract relevant data about PDB while constructing and specifically testing SMS components. Therefore, PDB_Mining is highly recommended for software developers while dealing with some of not best structured data from PDB. PDB_Mining provides following data: total residue occurrence in whole PDB, keyword search, PDB files sorted by file size, PDB files sorted by number of protein chains, PDB files sorted by X-ray resolution of 3D data, PDB files sorted by deposition date, PDB files classified with respect to number of DNA chains, protein families classification etc.
ConSSeq: displays the amino acid sequence of a given protein structure along with the consensus sequence (as found in HSSP), color coded with respect to the calculated degree of conservation. ConSSeq also displays color coded graphic bars of relative entropy as well as information about residues present in other homologous sequences, with their respective frequency. For fast visualization, this program also generates the sequence logo within very effective user interface (Fig. 3B).
Cα–Cα and Cβ–Cβ distance java plot: the Java Cα–Cα (Cβ–Cβ) distance plot is a diagram where the distances between the alpha (beta) carbon of one residue and all alpha (beta) carbon atoms of other residues, within a single chain of the PDB file, are represented by colored squares in a symmetrical plot (Fig. 3A). A user defined color code of those squares follows the scale of calculated distances among Cα–Cα or Cβ–Cβ atoms.
Protein Dossier (Fig. 2B): this SMS component provides a graphical summary of several important structural characteristics for a chosen protein. Protein Dossier displays the amino acid sequence cartoon, accompanied with two histograms representing the atomic contacts (as in the Graphical contacts module), as well as IFR residue identification and IFR contacts. Below the sequence cartoon there is a Prosite pattern information, followed by three different secondary structure indicators (PDB, DSSP, STRIDE). Another five parameters are displayed and annotated with color coded scales, representing for each amino acid the corresponding: temperature factor, sequence conservation in a multiple alignment (relative sequence entropy), solvent accessibility of the protein chain in isolation and in complex with other protein chain present in PDB file, and finally, dihedral angles. Protein Dossier presentation is designed so that it can justify the very name of this module. Protein Dossier is a tool that a user can employ to browse through valuable information about the protein structure. The collection of information presented (growing with each new SMS versions release) is allowing a user to make very knowledgeable decisions about the possible role of specific amino acids in defining the function of the protein. It also helps in deciphering what effect the specific mutation will have on the structure and function of the protein, specifically by observing changes in intra and interface contact signatures.
Figure 2.
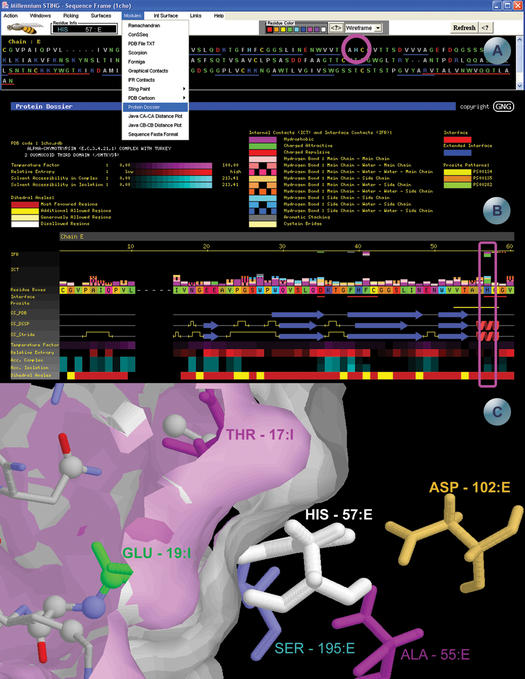
SMS example screens for the pdb file 1cho.pdb, highlighting the residue His57 in chain E. (A) Sequence frame, from where all the SMS modules and features can be accessed. The sequence is colored according to physico-chemical properties of the amino acids. The blue and red lines underneath the amino acid sequence represents the secondary structure elements (beta strands and helices respectively), according to the pdb file annotation. As the user scrolls the mouse over the sequence, information about the residues appears in the ‘residue info’ box. Two pull-down menus are also shown, demonstrating choice of SMS modules and rendering. (B) Protein Dossier module—the meaning of the colors are on the top part of the image. The internal and interface forming contacts are shown above the amino acid sequence cartoon, color coded according to the type of contact. The first row (red line) under the sequence highlights the residues at the interface. The next row (yellow line bellow the sequence stretch: VTAAHGC) indicates the PROSITE pattern PS00134. The following three rows indicate the secondary structure according to the annotations in PDB, DSSP and Stride, respectively. Another five rows are displayed color coded according to: temperature factor, relative entropy, accessibility in complex, accessibility in isolation and dihedral angles. His_57 and parameters belonging to this residue are placed within magenta quadrant. (C) 1cho structure representation rendered in the SMS structure window. The two surfaces represent the interface between the chain E (white) and chain I (pink). The figure was centered on the His57:E residue, painted in white. The other residues drawn in stick presentation are the residues that make contacts with the histidine, colored according to the contacts they make, corresponding to the first two lines of the Protein Dossier in (B). This particular image was obtained by combination of SMS actions, starting with the left mouse click on the Graphical IFR contacts java window, right above His_57 residue. Subsequently, some viewing arrangements are done in terms of removal of undesired information and then followed by surface building for the portion that makes part of the interface between the enzyme and inhibitor chains.
Figure 3.
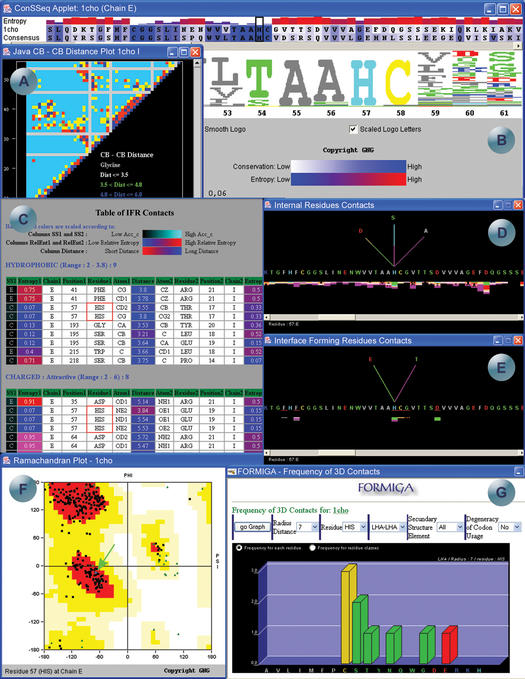
SMS example screens for the pdb file 1cho.pdb. (A) Java Cβ–Cβ distance plot, representing the distances between the beta carbons in chain I. (B) ConSSeq window where the amino acid sequence of 1cho, chain E is aligned with a consensus sequence as found in HSSP and, entropy histogram color and size coded with respect to the calculated degree of conservation. The size of sequence letters in the ‘logo’ follows the same degree of conservation, while color coding is the same as in SMS sequence frame. (C) Table of IFR contacts of the His_57, chain E amino acid. The table shows all the contacts in the protein, listing all the atoms along with its secondary structure, entropy, chain, residue number, residue name, atom name and distance in Å between two atoms in contact. The accessibility, entropy and distances of all atoms involved in the contacts are color coded and presented as background color. The accessibility is colored in the first column, ranging from black (low accessibility) to cyan (high accessibility), the entropy coloring scheme ranges from blue (low entropy) to red (high entropy) and the distances are colored from red (short) to blue (long). (D) Internal residue contacts formed by the His_57, chain E, showing all amino acids, belonging to the same chain E, that make contacts with it. The fan above the residue is color coded according to the type of contact the residue is involved with. (E) Interface forming residues contacts formed by the His_57, chain E. (F) Ramachandran plot of 1cho.pdb, highlighting the position of His_57, chain E in the diagram, and coloring plotted areas according to the allowed regions for the phi and psi angles. (G) Module FORMIGA showing the frequency of neighbors to the histidine residues (all His residues of E chain are considered and data demonstrated as a sum of calculated hits) found within the 7 Å radius sphere, centered at last heavy atom (LHA) of the histidine side chain.
STING Millennium FEATURES
The main aspect of the STING Millennium is its ability to combine data delivery through the web with the structural analysis tools, in order to provide a self-contained instrument for macromolecular studies. More than a simple front-end to the Chime plugin, SMS offers analytical services which we will only briefly describe here, counting that users will refer to extensive on-line help for further details.
STING Millennium is composed of two main windows. The SMS sequence window (Fig. 2A) displays sequence and contains the general menus with the commands and the SMS structure window (Fig. 2C) that renders the macromolecular 3D structure. In general terms STING Millennium provides the following services:
Ability to easily select residues in the sequence, select elements of secondary structure, as well as offer a wide variety of methods for rendering and coloring a molecule (mostly available through the ACTION menu).
Defining 3D neighbors to arbitrary selected residues.
Building surfaces of whole molecule or just of the IFR part of it.
Interactive Ramachandran plot, permitting rapid identification of residues in the disallowed regions and display of selected residues in the structure window.
Calculation of residue frequency within selected chain or on interface, as well as frequency of those residues filtered through chosen contact parameters.
Hydrogen bond net calculation with special attention given to the participation of water molecules.
Contacts definition and calculation for the whole molecule and/or interfaces.
Convenient 2D graphical presentation of parameters extracted from the 3D structure.
Display of sequence neighbors and calculation of relative sequence conservation for the family of homologous proteins.
SMS provides a great number of links to other web databases and programs. Such extensive cross-referencing and linking allows a user to have a most complete and integrated environment for sequence/structure/function analyses.
EXAMPLE OF STING Millennium APPLICATION
In Figures 2 and 3 we show a collage of snapshots produced by different SMS modules during a session that analyzed alpha-chymotrypsin (E chain) in complex with turkey ovomucoid third domain (I chain) of structure available in the PDB file 1cho.pdb. Specific attention in this example is given to the charged His residue (residue number 57) in the E chain. Analyzing information collected by SMS components and displayed in Figures 2 and 3, a user can infer many valuable conclusions about how important this amino acid is for protein stability, for protein function and for binding to inhibitor/substrate:
SMS Sequence and control window (Fig. 2A): sequence window displays linear protein and/or DNA sequence. Amino acids are color coded according to hydrophobicity and charged groups. The SMS sequence window also shows the numbering of the residues in the sequence, gaps in the PDB sequence, a chain identifier and ranges for all secondary structure elements as annotated in the original PDB file. Each residue in the sequence window is ‘clickable’, resulting in a presentation of its position in the SMS structure window (Fig. 2C). There are seven different display options in the menu bar situated just left from the Refresh button on SMS control panel: Wireframe, WS=Wireframe+spacefill, CPK, Ribbon, Backbone, Strands and Cartoon. Blue and red lines below the sequence are also ‘clickable’ resulting in a graphical ribbon presentation of the specified sequence region (red lines indicate helical region and blue lines indicate extended sheets region). Circled in magenta in Figure 2A is His_57 of the E chain.
Cβ–Cβ distance plot shown in Figure 3A, displays I chain data. In this plot a user is able to immediately identify glycine residues as they do not have Cβ atoms and their presence is evident from the existence of a light gray color vertical and horizontal lines (missing distance values due to non-existing Cβ atoms).
Figure 3B shows SMS ConSSeq output centered at His_57. A user may confirm both from ‘logo’ and histogram presentation, that the conservation of His_57 residue in the protein sequence is very high (even larger than for the neighboring Cys_58, which makes disulphide bond with Cys_42).
In Figure 3C and D, we show tabular information on amino acid IFR contacts and fan-like virtual internal-contact lines coming out from residue His_57 of chain E, respectively. Those virtual contact lines point to an amino acid that makes a particular contact (identified by the color of the line connecting it) with His_57. Information about the distance, atom partners and type of contact is provided in text format and if a particular residue is selected with the mouse click, zoomed image is displayed in the SMS structure window, showing the contacting 3D environment in details. The histogram along sequence of the chain E aids in rapid localization of critical residues, defined as those having larger than average number of contacts. Also, by color differentiation, a user may spot those amino acids having more energy-valuable contacts (e.g. electrostatic interactions).
Interface Forming Residues (IFR) Graphical Contacts are shown in Figure 3E. This inset demonstrates the same sequence of the E chain, but now a user can see IFR as red underlined sequence regions. Crucial difference from Figure 3D is that here contacts are counted only between residues belonging to different chains. A user can easily spot that chosen residue His_57 of chain E, make a couple of different contacts with the I chain residues E and T, and within its own E chain, with residues D, S and A. All details about IFR contacts are given in the table shown in Figure 3C, and in graphical form in Figure 2C.
Ramachandran plot is displaying the main-chain dihedral angles (φ and ψ), in Figure 3F. Green arrow points to the values of His_57, a position indicative of alpha helical secondary structure.
SMS Formiga provided graphs in Figure 3G. This graphical presentation shows which amino acids are found within 7 Å radius sphere, centered at the Last Heavy Atom (LHA) of the histidine residues. LHAs are defined as non-hydrogen side chain atoms which are most distant from the Cα atom. This information may also be transformed in cumulative frequencies for hydrophobic, polar and charged groups (one click away from demonstrated graph). Differences in frequencies are visually much easier to analyze than otherwise. Clearly, this information is valuable to define local environment for any amino acid and within any type of secondary structure. Variation in the size of the probing sphere allows for fine calibration of the potential function describing local residue environments.
Finally, Figure 2B shows a summary of the data calculated by a number of SMS programs and collected into SMS_DB. Now a user may analyze all pertinent parameters within a single visual area in Protein Dossier generated image. This is a very powerful tool which enables a quick estimate of the level of engagement for each amino acid within its own protein chain and functionally more importantly, in the mechanism of binding to substrate and/or inhibitor. Consequently, a molecular biologist is able now to make intelligent predictions about effects of mutations (previously judged as necessary) on both protein structure and protein function.
CONCLUSIONS
STING Millennium Suite is described here in terms of a solution that brings together a number of protein analysis tools at a single web server. Furthermore, a user is now able to download and install a whole package on its own platform and have network independent version running. We hope that for many laboratories, STING Millennium Suite will be able to decipher some of the hidden 3D parameters and aid in research. For some others, we hope that STING Millennium Suite will show itself as a valuable didactic tool. In number of occasions, STING Millennium Suite features were tailored so that we can make progress in our own research (28,29); occasionally, such tools were non-existent, inadequate and/or too tedious to perform by standard copy–paste–adapt procedures. We have developed STING Millennium Suite keeping in mind simplicity of presentation and also a necessity for simple and quick reproduction of informative graphs on printouts. Such procedures are explained in detail in the help manual.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported in part by the following grants: FAPESP 01/08895-0, FINEP 1945/01 and CNPq 521093/2001-5 (NV) and by NSF grant #DBI-9904841 to Barry Honig. The authors wish to express special thanks to Phil Bourne and Wolfgang Bluhm for their collaboration in testing and maintaining SMS at the PDB site. Special thanks are expressed to Barry Honig for his valuable insights and continuous interest in this project.
REFERENCES
- 1.Berman H.H., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laskovski R.A., Hutchinson,E.G., Michie,A.D., Wallace,A.C., Jones,M.L. and Thornton,J.M. (1997) PDBsum: a web-based database of summaries and analyses of all PDB structures. Trends Biochem. Sci., 22, 488–490. [DOI] [PubMed] [Google Scholar]
- 3.Hogue C.W. (1997) Cn3D: a new generation of three-dimensional molecular structure viewer. Trends Biochem. Sci., 22, 314–316. [DOI] [PubMed] [Google Scholar]
- 4.Neshich G., Togawa,R.C. and Honig,B. (1998) Sequence To and withIN graphics PDB Viewer (STING-PDB viewer). PDB Quart. News., July (electronic edition), 6. [Google Scholar]
- 5.Sander C. and Schneider,R. (1991) Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins, 9, 56–68. [DOI] [PubMed] [Google Scholar]
- 6.Schneider R., de Daruvar,A. and Sander,C. (1997) The HSSP database of protein structure-sequence alignments. Nucleic Acids Res., 25, 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneider R. and Sander,C. (1996) The HSSP database of protein structure-sequence alignments. Nucleic Acids Res., 24, 201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bucher P. and Bairoch,A. (1994) A generalized profile syntax for biomolecular sequences motifs and its function in automatic sequence interpretation. In Altman,R., Brutlag,D., Karp,P., Lathrop,R. and Searls,D. (eds), ISMB-94; Proceedings 2nd International Conference on Intelligent Systems for Molecular Biology. AAAIPress, Menlo Park, pp. 53–61. [PubMed] [Google Scholar]
- 9.Nicholls A., Sharp,K. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- 10.Tsodikov O.V., Record,M.T. Jr. and Sergeev,Y.V. (2002) A novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J. Comput. Chem., 23, 600–609. [DOI] [PubMed] [Google Scholar]
- 11.Sridharan S., Nicholls,A. and Honig,B. (1992) A new vertex algorithm to calculate solvent accessible surface areas. Biophys. J., 61, A174. [Google Scholar]
- 12.Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometric features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- 13.Frishman D. and Argos,P. (1995) Knowledge-based protein secondary structure assignment. Proteins, 23, 566–679. [DOI] [PubMed] [Google Scholar]
- 14.Radzicka A. and Wolfenden,R. (1988) Comparing the polarities of the amino-acids–side-chain distribution coefficients between the vapor-phase, cyclohexane, 1-octanol, and neutral aqueous-solution. Biochemistry, 27, 1664–1670. [Google Scholar]
- 15.Gattiker A., Gasteiger,E. and Bairoch,A. (2002) ScanProsite: a reference implementation of a PROSITE scanning tool. Appl. Bioinf., 1, 107–108. [PubMed] [Google Scholar]
- 16.Gourley D. and Totty,B. (2002) HTTP: The Definitive Guide. O'Reilly and Associates, Sebastopol, CA. [Google Scholar]
- 17.Raggett D., Lam,J., Alexander,I. and Kmiec,M. (1998) Raggett on HTML 4. Addison-Wesley, Boston, MA. [Google Scholar]
- 18.Flanagan D. (1998) Java Script: The Definitive Guide. O'Reilly and Associates, Sebastopol, CA. [Google Scholar]
- 19.Campione M. and Walrath,K. (1996) The Java Tutorial—Object Oriented Programming for the Internet. Addison-Wesley Pub. Co. [Google Scholar]
- 20.Galtier N., Gouy,M. and Gautier,C. (1996) SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci., 1, 543–548. [DOI] [PubMed] [Google Scholar]
- 21.Page R.D.M. (1996) TREEVIEW: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci., 12, 357–358. [DOI] [PubMed] [Google Scholar]
- 22.Shindyalov I. and Bourne,P. (1998) Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng., 11, 739–747. [DOI] [PubMed] [Google Scholar]
- 23.Yang A.-S. and Honig,B. (1999) Sequence to structure alignment in comparative modeling using PrISM. Proteins, (Suppl. 3), 66–72. [DOI] [PubMed] [Google Scholar]
- 24.Ramachandran G.N., Ramakrishnan,C. and Sasisekharan,V. (1963) Stereochemistry of polypeptide chain configurations. J. Mol. Biol., 7, 95–99. [DOI] [PubMed] [Google Scholar]
- 25.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.GCG. Genetics Computer Group, (1991) Program Manual for the GCG Package, Version 7, 575 Science Drive, Madison, Wisconsin, USA. [Google Scholar]
- 28.Caldas C.A., Coelho,V.P.C.V., Rigden,D., Neshich,G., Moro,A.M. and Brigido,M. (2000) Design and synthesis of germline-based hemi-humanized single chain Fv against the CD18 surface antigen. Protein Eng., 13, 353–360. [DOI] [PubMed] [Google Scholar]
- 29.Da Silva M.C.M., Grossi de Sá,M.F., Chrispeels,M.J., Togawa,R.C. and Neshich,G. (2000) Analysis of structural and physico-chemical parameters involved in the specificity of binding between alpha-amylases and their inhibitors. Protein Eng., 13, 167–177. [DOI] [PubMed] [Google Scholar]