Abstract
Analysis of multiple sequence alignments can generate important, testable hypotheses about the phylogenetic history and cellular function of genomic sequences. We describe the MultiPipMaker server, which aligns multiple, long genomic DNA sequences quickly and with good sensitivity (available at http://bio.cse.psu.edu/ since May 2001). Alignments are computed between a contiguous reference sequence and one or more secondary sequences, which can be finished or draft sequence. The outputs include a stacked set of percent identity plots, called a MultiPip, comparing the reference sequence with subsequent sequences, and a nucleotide-level multiple alignment. New tools are provided to search MultiPipMaker output for conserved matches to a user-specified pattern and for conserved matches to position weight matrices that describe transcription factor binding sites (singly and in clusters). We illustrate the use of MultiPipMaker to identify candidate regulatory regions in WNT2 and then demonstrate by transfection assays that they are functional. Analysis of the alignments also confirms the phylogenetic inference that horses are more closely related to cats than to cows.
INTRODUCTION
Comparative genomics exploits the conservation of functional genomic sequences due to purifying selection to predict important segments, such as coding regions and gene regulatory elements (1–3). The goal is to distinguish orthologous sequences conserved because of purifying selection from those that still align but are no longer functional. Making this distinction is complicated by variable levels of selection on individual functional elements and variation in the rates of evolutionary change both between phylogenetic lineages and within genomes (4–9). Analysis of whole-genome alignments between human and mouse allows this variability in evolutionary rates to be incorporated into predictions of function based on pairwise alignments (9–12). However, it is clear that additional genome sequences improve the reliability of predictions of functional genomic sequences (9,13), and thus alignments of more than two sequences are needed.
Multiple alignments of genomic sequences of single loci or gene clusters have long been used as a guide to functional regions. Within regulatory regions, sequence-specific protein-binding sites are frequently revealed as blocks (‘phylogenetic footprints’) with significantly less sequence change than surrounding regions (14–17). These phylogenetic footprints can be reliable guides to novel functional elements of enhancers or promoters (18–20). However, the optimal phylogenetic distance over which one can find regulatory elements remains unresolved.
As a preview of the types of information that can be gleaned from the increasing numbers of genomic sequences being determined, the NISC Comparative Sequencing Program has sequenced ∼1 Mb of homologous sequence from several mammalian species, chicken and three fish in several target regions (13). Initial analysis of the multiple alignment of the region including CFTR on human chromosome 7 revealed new insights into patterns of conservation and evolution, as expected. Importantly, this study also showed that the set of highly conserved regions (i.e. those likely to be under purifying selection) identified from the multiple alignment could not be duplicated by adjusting the stringency of scoring parameters for a pairwise comparison between human and mouse sequences. Clearly, the aligned multiple sequences include considerable additional information that is not in a pairwise alignment.
Thus it is highly desirable for investigators to have access to fast, reliable computational tools for aligning long (of the order of 1 Mb) segments of genomic DNA from multiple species, just as there are for pairwise alignments (21–23). In this paper we describe the MultiPipMaker server for aligning two or more sequences, for which all but the first sequence can be draft quality. The server is not only an extension of the PipMaker model (21) to allow alignments of more sequences, but it also generates a true multiple alignment. This alignment program was used in the analysis of the CFTR region by Thomas et al. (13).
METHODS
Multiple alignment
Our approach begins by preparing a crude multiple alignment from the pairwise alignments between the reference sequence and each of the secondary sequences, which are computed by the blastz program (24). An initial pairwise alignment is composed of a set of local alignments, each of which contains aligned nucleotides and internal gaps. The local alignments can overlap with each other, as illustrated in Figure 1A. These overlaps are removed by a pruning process (Fig. 1A), resulting in each nucleotide in the reference sequence being aligned to, at most, one nucleotide in the secondary sequence. The alignment resulting from stringing the pruned local alignments together then contains letters A, G, C and T, with interspersed gap characters of two kinds, indicating internal gaps (within the local alignments) and end-gaps. The end-gaps lie between aligned segments in a secondary sequence. They are not penalized, so that as the multiple alignment is built and subsequently refined, the contiguous piece of secondary sequence is permitted to freely ‘float’ back and forth in its row. Internal gaps are penalized in the usual manner, with both a gap-open and a gap-extend penalty. The multiple aligner uses the blastz substitution scores (25) and the ‘quasi-natural gap costs’ described in detail by Altschul (26), with a straightforward extension that appropriately handles end-gaps.
Figure 1.
Constructing a multiple alignment. (A) Constructing a row of the crude multiple alignment. One of the secondary sequences (e.g. sequence r) consists of two contigs. The pairwise alignments between the reference sequence and the two contigs are shown in a dot-plot format, in which the positions of each local alignment are plotted as a series of diagonal lines. For clarity, the four major local alignments are numbered and enclosed in shaded parallelograms. To construct a row in the crude multiple alignment, the local alignments are pruned so that each position in the reference sequence is aligned at most once. In this illustration, interval a-b is aligned to the reverse complement of B–A, b–c is aligned to B–C, c–d is aligned to C′–D, and e–g is aligned to E–G. This necessitates some pruning since some positions in the reference sequence are aligned more than once, e.g. the positions just before b. Extraneous matches to an improperly masked repetitive element around position f are discarded. Row r of the crude multiple alignment is constructed from the aligned intervals listed above. Gaps within a local pairwise alignment, say between a and b, result in ‘internal gaps’ in row r of the multiple alignment, which are penalized. A region between aligned segments (e.g. region z–a or d–e) is considered an ‘end-gap’ and is not penalized. Note that segment E–D of the secondary sequence appears twice in row r. (B) Refinement of the multiple alignment. One cycle of the refinement process is shown schematically. The crude multiple alignment is shown as a series of rows with thick lines representing strings of nucleotides; gaps are spaces in the rows. A subalignment between positions i and j is extracted and row r removed. The subalignment and row r are reduced by removing gaps as described in the Methods, and a new alignment is computed between the sequence in row r and the reduced subalignment (without row r). If this process improves the alignment score, then the new subalignment is spliced back into the large alignment. This process is repeated for all sub-regions where the alignment's columns have changed.
The crude multiple alignment is processed by an iterative refinement procedure. In adopting this strategy, we were inspired by the success of Anson and Myers (27), though our application involves much more flexible alignment scores. The following basic steps are repeated (Fig. 1B). We are given alignment column positions i and j, and a row index r in the multiple alignment. The column positions are such that between them, row r is spanned by a contiguous piece of sequence r (i.e. there are no end-gaps in this part of row r). We extract the subalignment covering columns i to j, then remove row r from the subalignment. The subalignment is further reduced by discarding any columns that contain only internal-gap or end-gap symbols and the segment of row r is reduced by removing any internal-gap symbols. Then an optimal alignment is computed between the ‘sequence’ whose entries are columns of the small multiple alignment and the sequence composed from the segment of row r. If these steps improve the score, then the new alignment segment is spliced into the large alignment; otherwise, the large alignment is not changed. For fixed column positions i and j, this process is applied to each row of the alignment, and if the score is seen to improve in some sub-region of columns i–j, then we recursively refine that sub-region.
Tools for analyzing multiple alignments
To locate regions within the multiple alignment satisfying certain conditions we wrote the programs subalign, multi_pat and tffind, which can be downloaded from the PipMaker site. All three programs scan a textual form of the multiple alignment that can be requested from MultiPipMaker. Subalign extracts the aligned sequence of all species within a range of coordinates. Multi_pat finds conserved regions that match a user-specified nucleotide pattern; for instance it can be used to find conserved patterns not represented by a weight matrix.
Tffind identifies matches to position weight matrices (PWMs) in conserved regions within any number of sequences in an alignment, searching both the forward and reverse complement strands. The program sequentially searches through each position of each sequence in the alignment, examining strings of the length of the pattern defined by the weight matrix (commonly 6 nt long). For each of these strings, a score is computed directly from the PWM. Tffind stores the location of the strings whose score exceeds a threshold and then determines if they localize to the same position in the multiple alignment.
A powerful tffind option identifies clusters of putative transcription factor binding sites (28) within a multiple sequence alignment based on similarity to PWMs. Options for the user include the choice of a pattern, either by factor name or consensus sequence, the distance between desired sites, the minimum number of sequences that must match the pattern, the cut-off value required for a hit and range of alignment columns to search. A user can exclude coding regions from the search and choose a database for PWMs, such as IMD (29) or TRANSFAC (30). Additionally, the program can search a single file for matches to PWMs, singly or in batch mode. Documentation for tffind is given at the PipMaker website.
RESULTS
Multiple alignment algorithm
The major challenge in developing MultiPipMaker was to design a foolproof, completely automatic and efficient method for simultaneous alignment of several sequences in the megabase size range. Moreover, we required that any sequence, other than the first, can be given in unordered and unoriented contigs. The construction of the multiple alignment by MultiPipMaker proceeds in two phases, described in detail in the Methods. Briefly, in the first phase the reference sequence is aligned individually with each secondary sequence, a crude multiple alignment is prepared from the pairwise alignments, and overlaps in the local pairwise alignments are removed (Fig. 1A). In the second phase, the crude multiple alignment is refined, building upon a strategy employed by Anson and Myers (27), to generate a true multiple alignment using rigorously defined multiple-alignment scores (Fig. 1B).
A MultiPip reveals functional regions at various phylogenetic distances
A MultiPip is a set of percent identity plots showing the positions and percent identities of gap-free segments of alignments of each secondary sequence with the primary (reference) sequence. These graphs depict the pairwise alignments before generating the crude multiple alignment. Because the first sequence serves as the reference sequence, annotations indicating the name, position and transcriptional orientation of the genes are relative to the coordinates in the first sequence, as are the icons that represent repetitive elements.
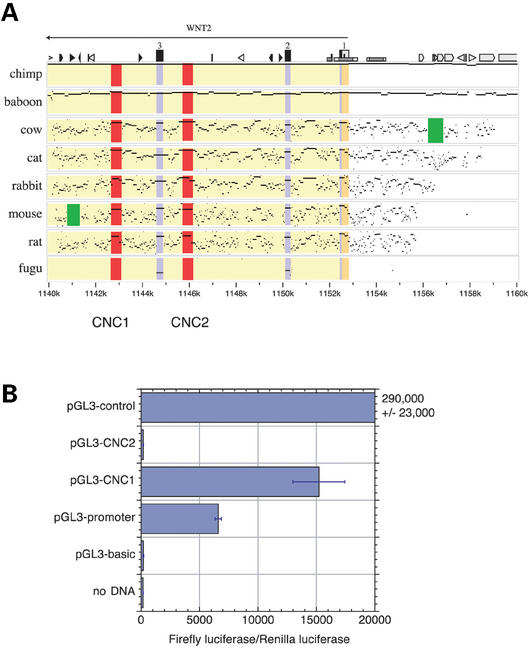
To illustrate features of a MultiPip, we use the subregion containing WNT2 within the larger 1.8 Mb region encompassing CFTR (13). WNT2 is one of a family of human homologs to the Drosophila segment polarity gene wingless. The WNT genes encode secreted glycoproteins that trigger a signal transduction pathway targeting beta-catenins and the Tcf/Lef family transcription factors. In addition, the WNT genes have been implicated in cell transformation (31). The MultiPip shown in Figure 2A provides a large-scale view of the pattern of matching sequences for the first three exons and introns of the WNT2 gene in human, chimp, baboon, cow, cat, rabbit, mouse, rat and pufferfish (Fugu). The panels are arranged roughly in order of increasing evolutionary distance from the human (reference) sequence. Both the amount of aligning sequence and the percent identity decrease with increasing distance, but the decrease is step-wise rather than smooth. All of the human sequence aligns with that of chimp, with very few mismatches, regardless of the type of sequence (coding, noncoding or repetitive) and an overall similarity of >98%. The comparison between human and baboon shows more mismatches and indels, but still most of the two sequences align. Comparisons with species in other mammalian orders show more mismatches and hence they can distinguish between coding and noncoding sequences. For instance, the exons begin to stand out in the human–cow comparison and they are distinguishable from most other sequences in the human–mouse and human–rat comparisons. In these latter comparisons, the coding exons are unbroken aligning segments of relatively high percent identity, whereas noncoding sequences either do not align or have many gaps in the alignment (32). The exons are clearly seen as ungapped alignments between human and fish, with very few alignments in noncoding regions for this gene. The WNT2 locus is more highly conserved than many other loci. Overall, for the entire 1.8 Mb region around CFTR, >90% of the human sequence aligns with sequences of other primates, ∼60% with those of carnivores and artiodactyls, ∼40% with those of rodents, and only ∼1% with that of fish, almost all of which is coding (13).
Figure 2.
Multiple percent identity plots (MultiPip) of the WNT2 region and tests of predicted regulatory elements. (A) MultiPip of the WNT2 region. Sequence data are from the June 2002 freeze of the NISC Comparative Sequencing Program (13). Local alignments between the human sequence and each second sequence (indicated on the left) are computed and displayed as the position in the human sequence (horizontal axis) and percent identity (from 50 to 100% along the vertical axis) of each gap-free aligning segment. Features in the human sequence are annotated above the graphs. Genes are labeled above arrows showing the direction of transcription, and exons are shown as numbered rectangles (black if protein-coding, gray if untranslated). Low rectangles denote CpG islands, shown as white if 0.6≤CpG/GpC<0.75 and as gray if CpG/GpC≥0.75. Interspersed repeats are shown by the following icons: white pointed boxes are L1 repeats, light gray triangles are SINEs other than MIR, black triangles are MIRs, black pointed boxes are LINE2s, and dark gray triangles and pointed boxes are other kinds of interspersed repeats, such as LTR elements and DNA transposons. Areas within these percent identity plots are colored light green for introns, blue for coding exons, yellow for noncoding exons, and red for notably conserved noncoding, nonrepetitive regions. Green boxes highlight lineage-specific deletions in cow and mouse. (B) Tests of CNCs for effects on expression after transient transfection. The indicated plasmids encoding firefly luciferase were transfected into HeLa cells in triplicate with a co-transfection control expressing Renilla luciferase. Test plasmids contained CNC1 or CNC2 inserted upstream of the SV40 promoter driving the luciferase gene. Enzyme activity in cell extracts was measured 48 h after transfection. The graph shows the means and standard errors of the activity ratios (firefly luciferase activity from the test plasmid divided by Renilla luciferase activity from the co-transfection control). Detailed methods are provided at the website http://bio.cse.psu.edu.
Prediction and confirmatory tests of gene regulatory sequences
Striking noncoding matches are seen flanking the third exon of WNT2 (Fig. 2A), which we call conserved noncoding sequences 1 and 2 (CNC1 and CNC2). The gap-free alignments between human and mouse in CNC1 and CNC2 are 418 and 439 bp, respectively, both at 87% identity. Hence they substantially exceed the criteria used by Loots et al. (33) to successfully predict a regulatory element. Furthermore, a multiple alignment conservation score that weights each contributing pairwise alignment by its evolutionary distance from human shows these as regions under stringent selection (13). They also score high for ‘regulatory potential’ based on human–mouse alignments analyzed by comparison to Markov models of patterns in the sequence alignments of known regulatory regions versus neutral DNA (12).
Given these strong computational predictions that CNC1 and CNC2 are functional and potentially regulatory, we tested their ability to affect the level of expression of a luciferase reporter gene driven by the SV40 enhancer (parental plasmid pGL3-promoter) after transient transfection into HeLa cells. Plasmids in which CNC1 was added to pGL3-promoter had a higher level of expression than the parental plasmid, whereas those containing CNC2 were effectively silenced (Fig. 2B). The negative effect of CNC2 was also seen after transient transfection in 293 cells (data not shown), in which the SV40 promoter is active independently of enhancers (34). These data indicate that both CNCs are cis-acting regulatory elements, with CNC1 having a modest positive effect and CNC2 being a potent silencer. Clearly, the regulatory elements predicted computationally can affect a heterologous promoter.
Analysis of a multiple alignment shows potential protein binding sites in predicted regulatory regions
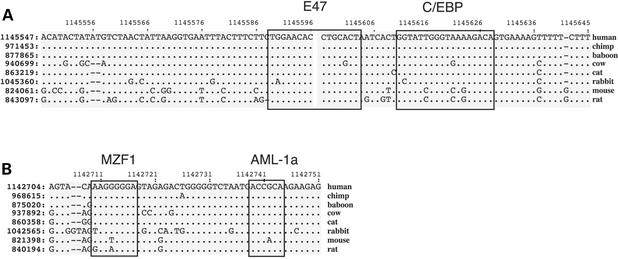
In addition to the aligned Pips shown in Figure 2, MultiPipMaker also computes a simultaneous alignment of the sequences. The multiple alignment is used for fine-structure, detailed analyses, including the identification of candidates for protein-binding sites. Matches of one sequence to PWMs describing binding sites for known transcription factors are found by programs such as MATINSPECTOR (35) or MATRIX SEARCH (29). We developed tools for finding candidate protein-binding sites in multiple alignments. The program tffind searches the multiple alignment for matches to PWMs describing such sites. Furthermore, tffind has a proximity feature, so that it identifies regions with conserved matches to one binding site that are within a user-specified distance from matches to a different binding site. When this program is applied to the multiple alignment of WNT2 CNCs, it locates conserved matches to PWMs for binding sites for E47 and C/EBP in CNC2 (Fig. 3A) and for MZF1 and AML-1a in CNC1 (Fig. 3B). When tffind searched the aligned sequences in the 1.8 Mb region surrounding CFTR (13) for blocks in which conserved binding sites for MZF1 and AML-1a were clustered within 100 bp, 10 were found, including the one at CNC1.
Figure 3.
Multiple alignments in the WNT2 CNCs annotated with matches to transcription factor binding sites. (A) Multiple alignment of part of CNC2 with a box drawn around the block identified by tffind as matching the E47-binding site. (B) Multiple alignment of part of CNC1 with boxes drawn around the blocks identified by tffind as matching the MZF1-binding site and the AML-1a-binding site.
Lineage-specific repeats indicate that horse and cat are sister species
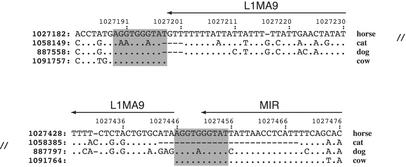
Multiple alignments are critical for phylogenetic analysis. One example of this was inspired by the recent conclusion, based on molecular divergence, that horses are phylogenetically closer to carnivores than to cows and other artiodactyls (36,37). We confirm this by a completely independent analysis examining patterns of transposition events. The MultiPipMaker alignments show the same transposon element inserted in horse, cat and dog, but not in cow. The L1MA9 element shown in Figure 4 occurs at orthologous positions, in the same orientation, and with the same target-site duplications in horse and the two carnivores, while the target-site sequence appears only once in cow. The L1MA9 subfamily of LINE1 interspersed repeats is believed to have been active around the time of the eutherian radiation (Table 6 in 9).
Figure 4.
An interspersed repeat that supports a phylogenetic reconstruction with horse closer to carnivores than to cow. The arrow points toward the A-rich 3′ tail of the transposon. The target-site duplication is shaded. Note that the AGGTGGGTAT at positions 1091764-1091773 in cow is aligned twice by MultiPipMaker.
The L1MA9 element was discovered by running a special-purpose program that analyzed alignments. We also used it to search for repeat elements that might support contradictory hypotheses (cat as an outgroup for horse and cow, or horse as an outgroup for cat and cow), but no such examples could be found. The program was originally used to discover six transposon elements that support the proposed early-eutherian split between an ancestor of primates and rodents and an ancestor of carnivores, artiodactyls and horse (data not shown).
DISCUSSION
The increasing availability of genome sequences creates a strong demand for tools to align them and to display the results in a compact, readily understood format with easy links to functional annotation. The percent identity plots generated by our pairwise alignment server (PipMaker, 21) show both positions and degree of similarity for aligning segments throughout two long genomic DNA sequences. We now have extended this capacity so that a series of genomic sequences can be aligned, each as a pairwise comparison with the first sequence. Furthermore, the resulting alignments are combined by an iterative-improvement strategy that yields a simultaneous nucleotide-level alignment of all the input sequences.
Several other programs are available for constructing multiple alignments of protein or DNA sequences, but none are fully suitable for our needs. Programs such as ClustalW (38) generate multiple alignments of protein sequences or relatively short genomic DNA sequences (reviewed in 39), but they are not designed to align long (megabase sized) DNA sequences. The VISTA server (40) will generate global alignments of multiple long DNA sequences. However, we require that any sequence, other than the first, can be given in unordered and unoriented contigs, and MultiPipMaker is the only available multiple alignment program designed for this task. Both the MultiPipMaker and the PipMaker (21) servers generate all local alignments above an appropriate threshold, which in principle can show every meaningful match among the sequences while allowing for duplications and inversions. Differences between the global and local approaches are discussed in Frazer et al. (23). A very recent addition to the arsenal of programs for aligning multiple genome sequences is described by Brudno et al. (41), who give additional background on the problem.
Gene regulatory elements can be identified as motifs over-represented in the upstream sequences of co-expressed genes in yeast, using programs such as AlignACE (42,43). The greater sequence complexity of mammals complicates the application of such techniques to single genomic DNA sequences; however, restricting the search to DNA segments conserved between human and mouse greatly enriches for known regulatory motifs and allows the computational determination of binding specificities (44). Indeed, strong conservation alone can be sufficient to predict regulatory elements from pairwise alignments (33), but the varying rate of evolution at different loci means that pairwise alignments are not adequate for identifying candidate regulatory elements at all loci (9,11). Newer predictive methods that take into account local rate variation, such as L-scores (9), should improve the effectiveness of pairwise alignments. The additional information in a multiple sequence alignment provides even greater resolution of highly conserved, likely functional sequences (13). We show that two particularly well-conserved intronic sequences in WNT2 can affect the level of reporter gene expression driven by the SV40 promoter. This effect on a heterologous promoter supports the hypothesis that these CNCs play a role in regulation of WNT2 gene expression. Combining computational results based on independent criteria, such as over-representation of motifs in co-expressed genes (42), clusters of matches to known transcription factor-binding sites (45), strong conservation (33), and similarity of patterns in alignments to those in known regulatory elements (12), should improve the accuracy of predictions of regulatory elements, especially as the algorithms are refined by experimental tests.
The evolutionary distance is determined both by phylogenetic distance (years since divergence) and by evolutionary rates within a lineage or at a particular locus. For instance, the higher rate of evolution in the rodent lineage (9) means that the human–rodent comparisons cover a greater evolutionary distance than human–carnivore comparisons, even though primates and rodents are sister taxa clearly separated from other eutherian orders by molecular criteria (13,37). Given the variation in evolutionary rates across mammalian genomes, the evolutionary distance at which particular features, such as coding exons or regulatory regions, begin to stand out in the MultiPip will differ from locus to locus. This is one of the powerful features of multiple alignments with a large number of species; it is difficult to know a priori which pairwise comparisons will be optimally informative and hence it is better to examine several comparison species. Consider the use of mammal–fish comparisons to predict exons. In the WNT2 locus examined here, many but not all exons are found by this pairwise comparison. Thus for some cases, mammal–fish comparisons are too stringent to find all exons, but examination of alignments to several species at a closer evolutionary distance will help identify the missed exons.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Blair Hedges for advice on mammalian phylogenetics. This work was supported by PHS grants HG02238 (to W.M.), DK27635 (to R.C.H.), and HG02325 (to L.E.). E.D.G. and the NISC Comparative Sequencing Program were supported by funds from the National Human Genome Research Institute. The following individuals were key contributors within the NISC Comparative Sequencing Program: Jim Thomas (BAC isolation and mapping); Jeff Touchman and Bob Blakesley (BAC sequencing); Gerry Bouffard, Steve Beckstrom-Sternberg, Pam Thomas, Jenny McDowell, Baishali Maskeri, Nancy Hansen, and Elliott Margulies (computational analyses).
REFERENCES
- 1.Kimura M. (1977) Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature, 267, 275–276. [DOI] [PubMed] [Google Scholar]
- 2.Li W.H., Gojobori,T. and Nei,M. (1981) Pseudogenes as a paradigm of neutral evolution. Nature, 292, 237–239. [DOI] [PubMed] [Google Scholar]
- 3.Pennacchio L.A. and Rubin,E.M. (2001) Genomic strategies to identify mammalian regulatory sequences. Nature Rev. Genet., 2, 100–109. [DOI] [PubMed] [Google Scholar]
- 4.Li W., Ellsworth,D., Krushkal,J., Chang,B. and Hewett-Emmett,D. (1996) Rates of nucleotide substitution in primates and rodents and the generation-time effect hypothesis. Mol. Phylogenet. Evol., 5, 182–187. [DOI] [PubMed] [Google Scholar]
- 5.Wolfe K.H., Sharp,P.M. and Li,W.H. (1989) Mutation rates differ among regions of the mammalian genome. Nature, 337, 283–285. [DOI] [PubMed] [Google Scholar]
- 6.Hardison R., Krane,D., Vandenbergh,D., Cheng,J.-F., Mansberger,J., Taddie,J., Schwartz,S., Huang,X. and Miller,W. (1991) Sequence and comparative analysis of the rabbit α-like globin gene cluster reveals a rapid mode of evolution in a G+C rich region of mammalian genomes. J. Mol. Biol., 222, 233–249. [DOI] [PubMed] [Google Scholar]
- 7.Hardison R., Oeltjen,J. and Miller,W. (1997) Long human-mouse sequence alignments reveal novel regulatory elements: A reason to sequence the mouse genome. Genome Res., 7, 959–966. [DOI] [PubMed] [Google Scholar]
- 8.Endrizzi M., Huang,S., Scharf,J.M., Kelter,A.R., Wirth,B., Kunkel,L.M., Miller,W. and Dietrich,W.F. (1999) Comparative sequence analysis of the mouse and human Lgn1/SMA interval. Genomics, 60, 137–151. [DOI] [PubMed] [Google Scholar]
- 9.International Mouse Genome Sequencing Consortium (2002) Initial sequencing and comparative analysis of the mouse genome. Nature, 420, 520–562. [DOI] [PubMed] [Google Scholar]
- 10.Li J. and Miller,W. (2002) Significance of interspecies matches when evolutionary rate varies. RECOMB 2002: Proceedings of the Sixth Annual International Conference on Computational Biology, ACM Press, New York, NY, pp. 216–224. [Google Scholar]
- 11.Hardison R.C., Roskin,K.M., Yang,S., Diekhans,M., Kent,W.J., Weber,R., Elnitski,L., Li,J., O'Connor,M., Kolbe,D. et al. (2003) Co-variation in frequencies of substitution, deletion, transposition and recombination during eutherian evolution. Genome Res., 13, 13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elnitski L., Hardison,R.C., Li,J., Yang,S., Kolbe,D., Eswara,P., O'Connor,M.J., Schwartz,S., Miller,W. and Chiaromonte,F. (2003) Distinguishing regulatory DNA from neutral sites. Genome Res., 13, 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas J.W. and Touchman,J.W. (2002) Vertebrate genome sequencing: building a backbone for comparative genomics. Trends Genet., 18, 104–108. [DOI] [PubMed] [Google Scholar]
- 14.Efstratiadis A., Posakony,J.W., Maniatis,T., Lawn,R.M., O'Connell,C., Spritz,R.A., DeRiel,J.K., Forget,B.G., Weissman,S.M., Slightom,J.L. et al. (1980) The structure and evolution of the human β-globin gene family. Cell, 21, 653–668. [DOI] [PubMed] [Google Scholar]
- 15.Tagle D.A., Koop,B.F., Goodman,M., Slightom,J., Hess,D.L. and Jones,R.T. (1988) Embryonic ɛ and γ globin genes of a prosimian primate (Galago crassicaudatus): Nucleotide and amino acid sequences, developmental regulation and phylogenetic footprints. J. Mol. Biol., 203, 7469–7480. [DOI] [PubMed] [Google Scholar]
- 16.Gumucio D., Shelton,D., Zhu,W., Millinoff,D., Gray,T., Bock,J., Slightom,J. and Goodman,M. (1996) Evolutionary strategies for the elucidation of cis and trans factors that regulate the developmental switching programs of the beta-like globin genes. Mol. Phylog. Evol., 5, 18–32. [DOI] [PubMed] [Google Scholar]
- 17.Hardison R., Slightom,J.L., Gumucio,D.L., Goodman,M., Stojanovic,N. and Miller,W. (1997) Locus control regions of mammalian β-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene, 205, 73–94. [DOI] [PubMed] [Google Scholar]
- 18.Elnitski L., Miller,W. and Hardison,R. (1997) Conserved E boxes function as part of the enhancer in hypersensitive site 2 of the β-globin locus control region: role of basic helix-loop-helix proteins. J. Biol. Chem., 272, 369–378. [DOI] [PubMed] [Google Scholar]
- 19.Shelton D.A., Stegman,L., Hardison,R., Miller,W., Slightom,J.L., Goodman,M. and Gumucio,D.L. (1997) Phylogenetic footprinting of hypersensitive site 3 of the β-globin locus control region. Blood, 89, 3457–3469. [PubMed] [Google Scholar]
- 20.Gottgens B., Barton,L.M., Gilbert,J.G., Bench,A.J., Sanchez,M.J., Bahn,S., Mistry,S., Grafham,D., McMurray,A., Vaudin,M. et al. (2000) Analysis of vertebrate SCL loci identifies conserved enhancers. Nat. Biotechnol., 18, 181–186. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz S., Zhang,Z., Frazer,K.A., Smit,A., Riemer,C., Bouck,J., Gibbs,R., Hardison,R. and Miller,W. (2000) PipMaker—a web server for aligning two genomic DNA sequences. Genome Res., 10, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayor C., Brudno,M., Schwartz,J.R., Poliakov,A., Rubin,E.M., Frazer,K.A., Pachter,L.S. and Dubchak,I. (2000) VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics, 16, 1046–1047. [DOI] [PubMed] [Google Scholar]
- 23.Frazer K.A., Elnitski,L., Church,D., Dubchak,I. and Hardison,R.C. (2003) Cross-species sequence comparisons: A review of methods and available resources. Genome Res., 13, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwartz S., Kent,W.J., Smit,A., Zhang,Z., Baertsch,R., Hardison,R.C., Haussler,D. and Miller,W. (2003) Human-mouse alignments with. Blastz. Genome Res., 13, 103–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiaromonte F., Yap,V.B. and Miller,W. (2002) Scoring pairwise genomic sequence alignments. Pac. Symp. Biocomput., 115–126. [DOI] [PubMed] [Google Scholar]
- 26.Altschul S.F. (1989) Gap costs for multiple sequence alignment. J. Theor. Biol., 138, 297–309. [DOI] [PubMed] [Google Scholar]
- 27.Anson E.L. and Myers,E.W. (1997) ReAligner: a program for refining DNA sequence multi-alignments. J. Comput. Biol., 4, 369–383. [DOI] [PubMed] [Google Scholar]
- 28.Mehldau G. and Myers,G. (1993) A system for pattern matching applications on biosequences. Comput. Appl. Biosci., 9, 299–314. [DOI] [PubMed] [Google Scholar]
- 29.Chen Q., Hertz,G. and Stormo,G. (1995) MATRIX SEARCH 1.0: a computer program that scans DNA sequences for transcriptional elements using a database of weight matrices. Comput. Appl. Biosci., 11, 563–566. [DOI] [PubMed] [Google Scholar]
- 30.Matys V., Fricke,E., Geffers,R., Gossling,E., Haubrock,M., Hehl,R., Hornischer,K., Karas,D., Kel,A.E., Kel-Margoulis,O.V. et al. (2003) TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res., 31, 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young C., Kitamura,M., Hardy,S. and Kitajewski,J. (1998) Wnt-1 induces growth, cytosolic beta-catenin, and Tcf/Lef transcriptional activation in Rat-1 fibroblasts. Mol. Cell. Biol., 18, 2474–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ansari-Lari M.A., Oeltjen,J.C., Schwartz,S., Zhang,Z., Muzny,D.M., Lu,J., Gorrell,J.H., Chinault,A.C., Belmont,J.W., Miller,W. et al. (1998) Comparative sequence analysis of a gene-rich cluster at human chromosome 12p13 and its syntenic region in mouse chromosome 6. Genome Res., 8, 29–40. [PubMed] [Google Scholar]
- 33.Loots G.G., Locksley,R.M., Blankespoor,C.M., Wang,Z.E., Miller,W., Rubin,E.M. and Frazer,K.A. (2000) Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science, 288, 136–140. [DOI] [PubMed] [Google Scholar]
- 34.Robbins P.D., Rio,D.C. and Botchan,M.R. (1986) Transactivation of the simian virus 40 enhancer. Mol. Cell. Biol., 6, 1283–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quandt K., Frech,K., Karas,H., Wingender,E. and Werner,T. (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res., 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar S. and Hedges,S.B. (1998) A molecular timescale for vertebrate evolution. Nature, 392, 917–920. [DOI] [PubMed] [Google Scholar]
- 37.Murphy W.J., Eizirik,E., O'Brien,S.J., Madsen,O., Scally,M., Douady,C.J., Teeling,E., Ryder,O.A., Stanhope,M.J., de Jong,W.W. et al. (2001) Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science, 294, 2348–2351. [DOI] [PubMed] [Google Scholar]
- 38.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson J.D., Plewniak,F. and Poch,O. (1999) A comprehensive comparison of multiple sequence alignment programs. Nucleic Acids Res., 27, 2682–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dubchak I., Mayor,C., Brudno,M., Pachter,L.S., Rubin,E.M. and Frazer,K.A. (2000) Active conservation of noncoding sequences revealed by 3-way species comparisons. Genome Res., 10, 1304–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brudno M., Do,C.B., Cooper,G.M., Kim,M.F., Davydov,E., NISC Comparative Sequencing Program, Green,E.D., Sidow,A. and Batzoglou,S. (2003) LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res., 13, 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roth F.P., Hughes,J.D., Estep,P.W. and Church,G.M. (1998) Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat. Biotechnol., 16, 939–945. [DOI] [PubMed] [Google Scholar]
- 43.Hughes J.D., Estep,P.W., Tavazoie,S. and Church,G.M. (2000) Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol., 296, 1205–1214. [DOI] [PubMed] [Google Scholar]
- 44.Wasserman W.W., Palumbo,M., Thompson,W., Fickett,J.W. and Lawrence,C.E. (2000) Human–mouse genome comparisons to locate regulatory sites. Nature Genet., 26, 225–228. [DOI] [PubMed] [Google Scholar]
- 45.Berman B.P., Nibu,Y., Pfeiffer,B.D., Tomancak,P., Celniker,S.E., Levine,M., Rubin,G.M. and Eisen,M.B. (2002) Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl Acad. Sci. USA, 99, 757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]