Abstract
We report the development of new software, OligoDesign, which provides optimal design of LNA (locked nucleic acid) substituted oligonucleotides for functional genomics applications. LNAs constitute a novel class of bicyclic RNA analogs having an exceptionally high affinity and specificity toward their complementary DNA and RNA target molecules. The OligoDesign software features recognition and filtering of the target sequence by genome-wide BLAST analysis in order to minimize cross-hybridization with non-target sequences. Furthermore it includes routines for prediction of melting temperature, self-annealing and secondary structure for LNA substituted oligonucleotides, as well as secondary structure prediction of the target nucleotide sequence. Individual scores for all these properties are calculated for each possible LNA oligonucleotide in the query gene and the OligoDesign program ranks the LNA capture probes according to a combined fuzzy logic score and finally returns the top scoring probes to the user in the output. We have successfully used the OligoDesign tool to design a Caenorhabditis elegans LNA oligonucleotide microarray, which allows monitoring of the expression of a set of 120 potential marker genes for a variety of stress and toxicological processes and toxicologically relevant pathways. The OligoDesign program is freely accessible at http://lnatools.com/.
INTRODUCTION
The challenge of creating comprehensive molecular catalogs of genes and proteins as well as establishing gene function and understanding biological processes for a specific organism demands the help of robust technologies, which can be applied to the exploration of a large number of parameters simultaneously. DNA chip technology utilizes miniaturized arrays of DNA molecules immobilized on solid surfaces for biochemical analyses. The power of DNA microarrays as experimental tools relies on the ultra specific molecular recognition of a specific element via complementary base pairing in combination with their miniaturized scale for the performance of massively parallel analyses. In the post-genomic era, microarrays have thus become the method of choice for many hybridization-based assays, such as expression profiling, DNA re-sequencing and genotyping on a genomic scale (1–7). Expression microarrays are capable of profiling gene expression patterns of tens of thousands of genes in a single experiment (8–10). On the other hand, the success of exploiting microarrays in functional genomics depends on continuous optimization of the array technology and the development of improved, cost-effective microarray platforms for producing accurate, reproducible and valid data.
DNA oligonucleotide microarrays have become increasingly popular in functional genomics applications. Several recent reports have focused on the sensitivity and specificity of different oligonucleotide-based microarray platforms demonstrating the usefulness of DNA oligonucleotide arrays in expression profiling compared to cDNA arrays (11–15). A common challenge for all DNA oligonucleotide microarrays is the need for gene specific oligonucleotides and an adequate compromise with respect to the sensitivity and specificity in the platform used. Recently, we have described a new microarray platform based on the use of locked nucleic acid (LNA) oligonucleotides, a novel class of bicyclic RNA analogs having an exceptionally high affinity toward their complementary DNA and RNA target molecules (16–18). Spotting of LNA-substituted oligonucleotides onto expression microarrays alongside the corresponding DNA oligonucleotides allowed direct comparison between the two probe types in expression profiling experiments. Our studies showed that introduction of LNA substitutions to DNA oligonucleotide capture probes resulted in a significant improvement in the discrimination between highly similar (90% sequence identity) mRNAs with a simultaneous increase in on-chip capture sensitivity (18). In addition, the use of LNAs allows full control of the melting temperature across microarray hybridizations (19). Since LNA chemistry is completely compatible with conventional DNA phosphoramidite chemistry, LNA substituted mixmer oligonucleotides can be designed to optimize performance.
The in silico design of optimal DNA oligonucleotides for functional genomics applications has been addressed previously by several non-commercial as well as commercial software packages (20–22; Array Designer: www.premierbiosoft.com/dnamicroarray/dnamicroarray.html; Pick70: se.osxgnu.org/pub/mirrors/sourceforge/arrayoligosel/; and Biosap: biosap.sourceforge.net/). Given the improved performance of LNA substituted oligonucleotides compared to DNA probes in expression profiling applications, it is desirable to develop general LNA design guidelines and automated software tools not presently available. In order to facilitate the use of LNA substituted oligonucleotides in functional genomics, we describe here a new software tool, designated OligoDesign, which addresses the key issues in the design of optimal LNA oligonucleotide capture probes for expression profiling microarrays. The OligoDesign program features several parameters, including recognition and filtering of the target sequence in the genome of interest, genome-wide BLAST analysis for minimized cross-hybridization, LNA melting temperature prediction, prediction of LNA self-annealing and secondary structure prediction for the LNA probe as well as the target nucleotide sequence. The optimal LNA substituted oligonucleotide for each gene is selected by applying fuzzy logic methods (23). Individual scores for the aforementioned parameters are calculated for each possible oligonucleotide in the query gene and run through a fuzzification process with decision functions adapted from the neural network community (24). Finally, the program ranks the LNA oligonucleotide probes according to the combined fuzzy logic score and returns the top scoring probes to the user in the resulting web page. The OligoDesign program is freely accessible at http://lnatools.com/.
THE OLIGODESIGN PROGRAM
General work flow
The OligoDesign program accepts nucleotide target sequences in FASTA format (25) as input and provides a web page with the predicted LNA substituted oligonucleotide capture probes and their properties as output. On the input page, a number of parameters guiding the oligonucleotide design can be adjusted. The resulting web page provides tab-separated tables that can be copied into a spreadsheet for further processing and oligonucleotide data storage. The web server calls a combination of Perl and C programs running on a Unix platform. These programs are described in detail below.
Sliding window BLAST analysis
After submission of the target nucleotide sequence to the program, OligoDesign carries out a genome-wide search with the mRNA sequence as input using blastn (26) against the selected genome database. The available databases are listed on the web server. The blastn parameters are: W 9 -e 100 -F F -S 3; the word length (default 9), the blastn expectation value (default 100) and the strand-to-search (default both) can be adjusted by the user. Blastn hits of 50 nt in length or longer with >98% sequence homology are considered to be the target gene and are filtered from the results. A window of the desired oligonucleotide length is moved across the mRNA sequence. For each window, which represents a potential LNA oligonucleotide capture probe, all matches to other sequences in the database are extracted from the blastn results and divided into two scores: the maximum number of matching nucleotides and the longest continuous match. The user can specify a threshold for both of these values as a percentage of the desired oligonucleotide length. The two values are determined and stored for all the possible oligonucleotides.
Prediction of duplex thermal stability and Tm equalization using LNA substitutions
The Tm is determined with a thermodynamic model based on the formula described in (27,28) and modified to predict the melting temperatures of DNA–LNA mixmers based on melting temperature measurements of >1400 DNA–LNA duplexes (for details, please see http://lna-tm.com). The Tm prediction tool has a standard deviation of 1.6 and 5.0°C for DNA and DNA–LNA mixmer oligonucleotides, respectively. The higher prediction error for LNA oligonucleotides is due to the more complex properties of these oligonucleotides, rather than lack of experimental data. The Tm prediction tool is freely accessible at http://lnatools.com/.
Self-annealing
Self-annealing of the oligonucleotides is determined using the Smith–Waterman (29) sequence alignment algorithm. The match values of the scoring matrix have been determined by using the coefficients of linear models determined from in-house oligonucleotide Tm measurements (data not shown), while the mismatch values are based on experiments described by Koshkin et al. (30). The algorithm corresponds to a simple linear melting temperature prediction, when applied to perfectly matched oligonucleotides. A comparison of the self-annealing scores calculated using the matrix described here, with Tm values predicted by the Tm prediction tool revealed a prediction error of ∼10°C for DNA–LNA mixmers, which is only a few degrees higher than that achievable by the more complex algorithm of the Tm prediction tool.
Prediction of the secondary structures
The secondary structure prediction is carried out using the Nussinov algorithm (31), according to the same scoring matrix as used for the self-annealing. As above, the score gives an indication of the expected stability of the secondary structure. The self-annealing and secondary structure prediction tools are available at http://lnatools.com/. The secondary structures are predicted and filtered by calculating the most stable secondary structure within a 100 nt window moved across the target gene. The motivation for using a sliding window is that the fluorochrome-labelled target in expression microarray hybridizations is either fragmented (cRNA) or often not of full-length (cDNA), thereby creating a need for local secondary structure prediction. The number of nucleotides involved in a given predicted secondary structure is calculated.
Selection of the optimal LNA oligonucleotides
All parameters are calculated for every LNA oligonucleotide within the target nucleotide sequence. To determine how well an oligonucleotide fits the desired specifications, a fuzzification of the values is performed. The fuzzification formula is: 1/(1+ exp(−k(c−x)); where k is the slope, c the cut-off, and x is the value of the property under consideration. It is inspired by the decision function of neural networks (24) and is used to make a soft decision about how well a given parameter fits the oligonucleotide criteria entered by the user. The scores of all the oligonucleotide properties are multiplied to give the final score. The user can weight a specific parameter by adjusting the given value from 0 to 5; the higher the value the more heavily the parameter is weighed in the combined oligonucleotide score. If the final score is close to one, all properties are well above the desired criteria, if it is close to zero, one or more of the parameters failed to meet the criteria set by the user.
RESULTS AND DISCUSSION
DNA oligonucleotide microarrays have become increasingly popular in functional genomics applications. Several recent reports have demonstrated the usefulness of DNA oligonucleotide arrays in expression profiling (11–15). Compared to PCR products amplified from cDNA libraries, gene-specific oligonucleotides of 40–70 nt in length can be designed with optimized hybridization properties and minimized cross-hybridization. Besides controlled specificity, oligonucleotide microarrays enable detection of alternatively spliced mRNAs and discrimination between highly homologous transcripts. Furthermore, the use of oligonucleotides provides a more cost-effective platform avoid from clone tracking, PCR amplification and sequence verification associated with the fabrication of cDNA arrays. A common challenge for all DNA oligonucleotide microarrays is the need for adequate design of gene-specific oligonucleotides with optimal sensitivity. A unique oligonucleotide capture probe will ideally hybridize to its target only, without cross-hybridization to cDNAs representing other genes in the genome, while simultaneously being capable of detecting low mRNA levels in the cell. Potential cross-hybridization to similar sequences is typically detected by general purpose sequence search methods such as BLAST or FASTA (25,26), whereas secondary structure prediction and self-annealing of the oligonucleotide can be modelled with sequence alignment or fold methods (32,33). The melting temperature of the oligonucleotide must allow formation of a stable duplex with its target under the hybridization conditions used. Furthermore, the melting temperatures of the capture probes spotted onto a microarray must be normalized within a narrow Tm range enabling uniform hybridization conditions across the array. Other important oligonucleotide selection considerations are sequence complexity, position in the target gene and hybridization accessibility in the target. Sequence complexity can be evaluated as stretches of identical nucleotides. It is often an advantage to bias the oligonucleotide location close to the 3′ end of the mRNA due to the oligo(dT) priming in the cDNA target synthesis. On the other hand, stable secondary structures in the target can make it inaccessible for hybridization. Consequently, oligonucleotides localized in these regions should be filtered out from the selected oligonucleotide set. It is often not possible to design an oligonucleotide that exhibits all the desired properties, thus creating a need for convenient design programs capable of selecting the most optimal oligonucleotides. This requires careful weighing of all the selection criteria and possibly accepting an oligonucleotide even though some of its properties are suboptimal.
We have previously described the use of LNA oligonucleotides in highly accurate genotyping assays for the apolipoprotein B R3500G SNP (34), and the apolipoprotein E codon 112 and 158 SNPs (35), as well as multiplex genotyping of 20 SNPs implicated within the dysmetabolic syndrome and with the maturity onset diabetes of the young (MODY) (19). In a recent study, we demonstrated that allele-specific LNA probes facilitate accurate genotyping of human populations by direct competitive hybridization to microarrays of immobilized patient amplicons. In addition, we showed that LNA oligonucleotide microarrays are more sensitive and specific in gene expression profiling when compared to DNA oligonucleotide arrays, enabling discrimination between highly homologous mRNAs with a simultaneous increase in sensitivity (18).
In order to facilitate the design of optimal LNA substituted oligonucleotides, we have developed a Tm prediction tool, based on determination of the thermal stability of >1400 LNA–DNA oligonucleotide duplexes by UV spectroscopy, as well as a software tool, OligoDesign, for in silico design of LNA oligonucleotides for functional genomics. The OligoDesign program addresses the key issues in the design of LNA oligonucleotide capture probes for expression microarray applications by providing unique, gene-specific oligonucleotide capture probes of a given length, typically 40–80mers, with equalized melting temperature, minimized cross-hybridization with homologous genes, as well as minimized self-annealing and secondary structures. These parameters are determined for each possible capture oligonucleotide for the query target sequence and presented to a fuzzy logic scoring system. The probes are hereafter ranked according to the fuzzy logic prediction and the top scoring suggestions are returned.
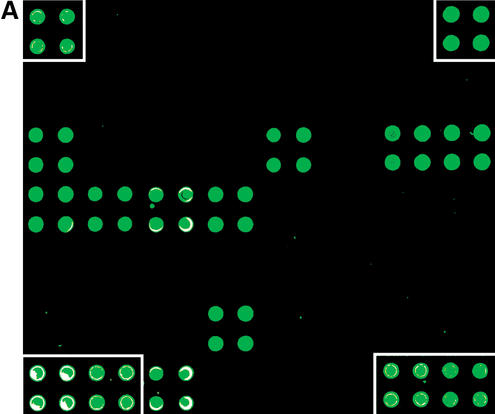
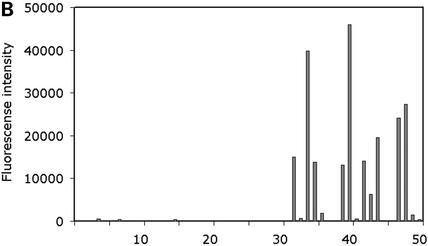
To demonstrate the utility of the OligoDesign program in optimal LNA oligonucleotide selection for expression profiling we have reported on the design and use of a Caenorhabditis elegans LNA tox microarray (18). The LNA oligonucleotide microarray monitors the expession of a set of 120 potential marker genes for a variety of stress and toxicity processes and toxicologically relevant pathways, including drug metabolism, DNA damage-repair, apoptosis, stress response, membrane proteins and cell cycle regulators. Gene-specific, 50mer LNA substituted oligonucleotides (with LNA at every third nucleotide position) were designed using the OligoDesign program, including capture probes for a selection of 25 cytochrome P450 genes with two oligonucleotides for each CYP450 target mRNA. The LNA oligonucleotides were synthesized with an anthraquinone group at the 5′ end enabling photocoupling of the capture probes onto polymer microarray slides. The specificity of the CYP450 LNA oligonucleotides was validated by 50 individual microarray hybridizations using biotin-labelled antisense oligonucleotides as targets for each CYP450 LNA oligonucleotide, followed by development of the hybridization signals using Cy3-labelled streptavidin. A strong, capture probe-specific hybridization signal was obtained from 47 CYP450 oligonucleotides with a signal-to-noise ratio of >5-fold compared to the probe showing highest cross-hybridization and an average signal-to-noise ratio of 230-fold across all CYP450 oligonucleotides. As an example, Figure 1 shows the microarray hybridization results from assessment of the specificity of the C.elegans CYP450 LNA oligonucleotide capture probes using a pool of 10 antisense target oligonucleotides. Two CYP450 oligonucleotides showed high cross-hybridization signals to each other due to the high similarity (80%) between the genes, which did not allow the OligoDesign program to design truly unique 50mer LNA capture probes for the two CYP450 target genes. Finally, one LNA capture probe was filtered from the validation data set due to a mislabeled antisense oligonucleotide. In addition to validation of the CYP450 capture probes, the LNA tox array has been used to investigate the transcriptional response to heat shock in developmentally staged C.elegans worms (18). Two heat shock proteins, HSP-70 (F44E5.4/5) and HSP-70 (H26D21.1) were found to be up-regulated in two independent microarray experiments, in accordance with previous C.elegans studies (36). Combined, these results imply that the OligoDesign program is highly useful in the design of unique LNA oligonucleotide capture probes for expression profiling microarrays.
Figure 1.
(A) Hybridization of the C.elegans cytochrome P450 LNA oligonucleotide microarray with a pool of 10 biotin-labelled antisense oligonucleotides. The 50 gene-specific CYP450 50mer LNA oligonucleotides were spotted in quadruplicate along with Cy3 labelled marker oligonucleotides (enclosed in white boxes) onto the Immobilizer microarray slide (Exiqon, Denmark). The resulting microarray was hybridized with a pool of 10 5′-biotin-labelled 50mer antisense target oligonucleotides at 65°C in 0.5 M NaCl, followed by staining with Streptavidin-Cy3 and scanning in a confocal laser scanner (ScanArray Express HT, Perkin Elmer, USA). A specific hybridization pattern is obtained from the 10 CYP450 LNA capture probes, whereas no cross-hybridization is observed for the remaining 40 CYP450 oligonucleotides. (B) Assessment of the specificity of the C.elegans CYP450 LNA oligonucleotide capture probes. Processing of the microarray data (GenePix Pro 4.01, Axon, USA) revealed strong capture probe-specific hybridization signals for the 10 CYP450 LNA oligonucleotides with a several fold higher signal intensity compared to the probe showing highest cross-hybridization.
In summary, we have developed a new software tool, which provides optimal in silico design of LNA substituted oligonucleotides for gene expression profiling. Given the high affinity of LNA oligonucleotides toward their complementary DNA and RNA targets together with their significantly improved discriminatory power, it would be highly attractive to integrate LNAs as enhancers to a wide variety of genomics applications. The OligoDesign software provides the researchers with the ability to design LNA oligonucleotides with optimized performance in nucleic acid recognition assays. The improved sensitivity and specificity of the LNA microarray platform may be especially advantageous in expression profiling of non-coding RNAs and assessment of alternative splicing.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Kenneth Harlow for critical reading of the manuscript and Janni J. Jørgensen for skilful technical assistance.
REFERENCES
- 1.Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 2.Lockhart D.J., Dong,H., Byrne,M.C., Follettie,M.T., Gallo,M.V., Chee,M.S., Mittmann,M., Wang,C., Kobayashi,M., Horton,H. and Brown,E.L. (1996) Expression monitoring by hybridisation to high-density oligonucleotide arrays. Nat. Biotechnol., 14, 1675–1680. [DOI] [PubMed] [Google Scholar]
- 3.Schena M., Shalon,D., Heller,R., Chai,A., Brown,P.O. and Davis,R.W. (1996) Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc. Natl Acad. Sci. USA, 93, 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shalon D., Smith,S.J. and Brown,P.O. (1996) A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res., 6, 639–645. [DOI] [PubMed] [Google Scholar]
- 5.DeRisi J.L., Iyer,V.R. and Brown,P.O. (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science, 278, 680–686. [DOI] [PubMed] [Google Scholar]
- 6.Eisen M.B. and Brown,P.O. (1999) DNA arrays for analysis of gene expression. Methods Enzymol., 303, 179–205. [DOI] [PubMed] [Google Scholar]
- 7.Lipshutz R.J., Fodor,S.P., Gingeras,T.R., Lockhart,D.J. (1999) High density synthetic oligonucleotide arrays. Nature Genet., 21, 20–24. [DOI] [PubMed] [Google Scholar]
- 8.Petricoin E.F. III, Hackett,J.L., Lesko,L.J., Puri,R.K., Gutman,S.I., Chumakov,K., Woodcock,J., Feigal,D.W.Jr, Zoon,K.C. and Sistare,F.D. (2002) Medical applications of microarray technologies: a regulatory science perspective. Nature Genet., 32, 474–479. [DOI] [PubMed] [Google Scholar]
- 9.Gerhold D., Lu,M., Xu,J., Austin,C., Caskey,C.T. and Rushmore,T. (2001) Monitoring expression of genes involved in drug metabolism and toxicology using DNA microarrays. Physiol. Genomics, 5, 161–170. [DOI] [PubMed] [Google Scholar]
- 10.Waring J.F., Jolly,R.A., Ciurlionis,R., Lum,P.Y., Praestgaard,J.T., Morfitt,D.C., Buratto,B., Roberts,C., Schadt,E. and Ulrich,R.G. (2001) Clustering of hepatotoxins based on mechanism of toxicity using gene expression profiles. Toxicol. Appl. Pharmacol., 175, 28–42. [DOI] [PubMed] [Google Scholar]
- 11.Kane M.D., Jatkoe,T.A., Stumpf,C.R., Lu,J., Thomas,J.D. and Madore,S.J. (2000) Assessment of the sensitivity and specificity of oligonucleotide (50mer) Microarrays. Nucleic Acids Res., 28, 4552–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai H., Meyer,M., Stepaniants,S., Ziman,M. and Stoughton,R. (2002) Use of hybridisation kinetics for differentiating specific from non-specific binding to oligonucleotide microarrays. Nucleic Acids Res., 30, e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramakrishnan R., Dorris,D., Lublinsky,A., Nguyen,A., Domanus,M., Prokhorova,A., Gieser,L., Touma,E., Lockner,R., Tata,M. et al. (2002) An assessment of Motorola CodeLink microarray performance for gene expression profiling applications. Nucleic Acids Res., 30, e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Relogio A., Schwager,C., Richter,A., Ansorge,W. and Valcarcel,J. (2002) Optimization of oligonucleotide-based DNA microarrays. Nucleic Acids Res., 30, e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H-Y., Malek,R.L., Kwitek,A.E., Greene,A.S., Luu,T.V., Behbahani,B., Frank,B., Quackenbush,J. and Lee,N.H. (2003) Assessing unmodified 70-mer oligonucleotide probe performance on glass-slide microarrays. Genome Biol., 4, R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koshkin A., Singh,S.K., Nielsen,P., Rajwanshi,V.K., Kumar,R., Meldgaard,M., Olsen,C.E., and Wengel,J. (1998) LNA (locked nucleic acids): synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron, 54, 3607–3630. [Google Scholar]
- 17.Singh S.K., Nielsen,P., Koshkin,A. and Wengel,J. (1998) LNA (locked nucleic acids): Synthesis and high-affinity nucleic acid recognition. Chem. Commun., 4, 455–456. [Google Scholar]
- 18.Kauppinen S., Nielsen,P.S., Mouritzen,P., Nielsen,A.T., Vissing,H., Møller,S. and Ramsing,N.B. (2003) LNA microarrays in genomics, PharmaGenomics, in press. [Google Scholar]
- 19.Mouritzen P., Nielsen,A.T., Pfundheller,H.M., Choleva,Y., Kongsbak,L. and Møller,S. (2003) SNP genotyping using LNA (Locked Nucleic Acid). Expert Rev. Mol. Diagn., 3, 89–100. [DOI] [PubMed] [Google Scholar]
- 20.Chen H., and Sharp,B.M. (2002) Oliz, a suite of Perl scripts that assist in the design of microarrays using 50mer oligonucleotides from the 3′ untranslated region. BMC Bioinformatics, 3, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rouillard J.M., Herbert,C.J. and Zuker,M. (2002) OligoArray: genome-scale oligonucleotide design for microarrays. Bioinformatics, 18, 486–487. [DOI] [PubMed] [Google Scholar]
- 22.Li F. and Stormo,G.D. (2001) Selection of optimal DNA oligos for gene expression arrays. Bioinformatics, 17, 1067–1076. [DOI] [PubMed] [Google Scholar]
- 23.Cox E. (1994) The Fuzzy Systems Handbook: A Practitioner's Guide to Building, Using, and Maintaining Fuzzy Systems. Academic Press, Boston. [Google Scholar]
- 24.Herz J., Krogh,A. and Palmer,R.G. (1991) Introduction to the Theory of Neural Computation. Addison Wesley, Reading. [Google Scholar]
- 25.Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.SantaLucia J. Jr (1998) A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA, 95, 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allawi H.T. and SantaLucia,J.Jr (1997) Thermodynamics and NMR of internal G:T mismatches in DNA. Biochemistry, 36, 10581–10594. [DOI] [PubMed] [Google Scholar]
- 29.Smith T.F., Waterman,M.S. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
- 30.Koshkin A.A., Nielsen,P., Meldgaard,M., Rajwanshi,V.K., Singh,S.K. and Wengel,J. (1998) LNA (Locked Nucleic Acid): an RNA mimic forming exceedingly stable LNA:LNA duplexes. J. Am. Chem. Soc., 120, 13252–13253. [Google Scholar]
- 31.Nussinov R., Shapiro,B., Le,S. and Maizel,J.V. (1990) Speeding up the dynamic algorithm for planar RNA folding. Math. Biosci., 100, 33–47. [DOI] [PubMed] [Google Scholar]
- 32.Zuker M., Mathews,D.H. and Turner,D.H. (1999) Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In Barciszewski,J. and Clark,B.F.C. (eds), RNA Biochemistry and Biotechnology, NATO ASI Series, Kluwer Academic Publishers, Dordrecht, The Netherlands, pp. 11–43. [Google Scholar]
- 33.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 34.Jacobsen N., Fenger,M., Bentzen,J. Rasmussen,S.L., Jakobsen,M.H., Fenstholt,J. and Skouv,J. (2002) Genotyping of the apolipoprotein B R3500Q mutation using immobilized locked nucleic acid capture probes. Clin. Chem., 48, 657–660. [PubMed] [Google Scholar]
- 35.Jacobsen N., Bentzen,J., Meldgaard,M., Jakobsen,M.H., Fenger,M., Kauppinen,S. and Skouv,J. (2002) LNA-enhanced detection of single nucleotide polymorphism in the apolipoprotein E. Nucleic Acids Res., 30, e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.GuhaThakurta D., Palomar,L., Stormo,G.D., Tedesco,P., Johnson,T.E., Walker,D.W., Lithgow,G., Kim,S. and Link,C.D. (2002) Identification of a novel cis-regulatory element involved in the heat shock response in Caenorhabditis elegans using microarray gene expression and computational methods. Genome Res., 12, 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]