Abstract
We have previously identified a Saccharomyces cerevisiae mutant that is markedly more resistant than wild-type to Dahlia merckii antimicrobial peptide 1 (DmAMP1), an antifungal plant defensin isolated from seeds of dahlia (Dahlia merckii). A complementation approach was followed that consisted of the introduction of a genomic library of DmAMP1-sensitive wild-type yeast into the DmAMP1-resistant yeast mutant and screening for restored sensitivity to DmAMP1. The gene determining sensitivity of S. cerevisiae to DmAMP1 was identified as IPT1, a gene encoding an enzyme involved in the last step of the synthesis of the sphingolipid mannose-(inositol-phosphate)2-ceramide. Strains with a nonfunctional IPT1 allele lacked mannose-(inositol-phosphate)2-ceramide in their plasma membranes, bound significantly less DmAMP1 compared with wild-type strains, and were highly resistant to DmAMP1-mediated membrane permeabilization. All of these phenotypic deviations could be restored by reintroduction of a functional IPT1 gene. Our data support a model in which membrane patches containing sphingolipids act as binding sites for DmAMP1 or, alternatively, are required to anchor membrane or cell wall-associated proteins, which themselves interact with DmAMP1.
Most, if not all, higher eukaryotes produce antimicrobial peptides that are thought to play a role in innate immunity mechanisms directed at keeping in check the growth of microorganisms inside their tissues (1). The antimicrobial peptides produced by different organisms vary widely in length, amino acid composition, and folding pattern. Many of them adopt α-helical structures in a lipophilic environment, whereas others, often those rich in disulfide-linked cysteines, have structures involving β-sheets. In nearly all cases studied so far, antimicrobial peptides have been found to affect the growth of microorganisms by means of interaction with membranes. For many peptides such interaction is mediated by common membrane phosphoglycerolipids such as phosphatidylserine or phosphatidylethanolamine. However, as some peptides exhibit strong specificity for particular types of microorganisms, it has often been suggested that the interaction between membranes and such peptides involves a specific docking site (1, 2), although no definitive proof has yet been presented for any of the eukaryotic antimicrobial peptides.
Plant defensins are small (45–54 amino acids), basic peptides that inhibit the growth of a broad range of fungi at micromolar concentrations (3, 4). These peptides share a characteristic three-dimensional folding pattern with an α-helix and a triple-stranded β-sheet, stabilized by eight disulfide-linked cysteines. On the basis of amino acid sequence homologies, plant defensins can be divided into several groups and subfamilies, which differ in their specificity for particular fungi (3, 5, 6). One of these subfamilies, subfamily A2, contains plant defensins such as Dahlia merckii antimicrobial peptide 1 (DmAMP1), AhAMP1 from Aesculus hippocastanum, and CtAMP1 from Clitoria ternatea, which share about 70% sequence identity with each other. Treatment of susceptible fungi with these plant defensins results in reduced hyphal elongation without marked morphological distortions (6).
A number of studies have attempted to unravel the mode of action of these plant defensins. It has been shown that DmAMP1 induces an array of relatively rapid responses in fungal membranes, including increased K+ efflux, increased Ca2+ uptake, increased uptake of fluorescent dyes, and membrane potential changes (7, 8). Furthermore, the existence of high-affinity binding sites for DmAMP1 on fungal cells and plasma membrane fractions was demonstrated (2). Binding of DmAMP1 to these sites was found to be irreversible, yet highly specific, as binding could not be competed for by more distantly related plant defensins, such as those belonging to subfamilies A3 and A4 (2). Two Saccharomyces cerevisiae mutants, DM1 and DM2, have been selected that exhibit increased resistance to DmAMP1, as well as to the related plant defensins AhAMP1 and CtAMP1. Plasma membrane fractions derived from these DmAMP1-resistant mutants have a severely reduced binding capacity for DmAMP1 (2). In addition, DmAMP1-mediated membrane permeabilization in these mutants is strongly reduced relative to wild-type strains (8). Hence it appears that membrane binding, membrane permeabilization, and growth inhibition caused by DmAMP1 are linked events.
Putting all of these observations together, we propose the following model for the mode of action of plant defensins. The first step in the path leading to fungal growth inhibition would be the binding of plant defensins to specific sites on the plasma membrane of fungal hyphae. Interaction with these binding sites subsequently would enable plant defensins to insert into the plasma membrane, thus affecting membrane structure and permeability to certain solutes, such as Ca2+ and K+, some of which play an important role in fungal growth and development.
In this paper we present evidence that a single gene from S. cerevisiae, namely the IPT1 gene, determines membrane binding, membrane permeabilization, and growth inhibition by the plant defensin DmAMP1. The gene product of the IPT1 gene catalyzes the conversion of mannose-(inositol-phosphate)-ceramide (MIPC) into mannose-(inositol-phosphate)2-ceramide [M(IP)2C] (9), the major sphingolipid in membranes of S. cerevisiae. We propose a model for the mode of action of plant defensins, in which membrane patches containing M(IP)2C constitute binding sites for DmAMP1 or, alternatively, are required for anchoring of membrane or cell-wall-associated proteins, which themselves interact with DmAMP1.
Materials and Methods
Strains.
S. cerevisiae strains used in this study are wild-type strain W303-1A (MATa leu2-3/112 ura3-1 trp1-1 his3-11/15 ade2-1 can1-100 GAL SUC2 IPT1 SUR1), wild-type strain YPH250 (10), and the corresponding ipt1-deletion mutant RCD113 (9). DmAMP1-resistant yeast mutants DM1 and DM2, which are derived from wild-type strain W303-1A, have been described previously by Thevissen et al. (2). Their relevant genotypes are DM1 (ipt1W93amber) and DM2 (ipt1Q227K). W303-1A derivatives DM3 (ipt1L268F) and DM4 (ipt1S61R) have been generated in this study. The sur1Δ strains W303-1AΔsur1 and DM1Δsur1 were constructed by disrupting SUR1 in W303-1A and DM1, respectively, as described below.
Antifungal Activity Assay.
Antifungal activity of protein samples against S. cerevisiae was assayed by microspectrophotometry of liquid cultures grown in microtiter plates as described previously (2, 8, 11). Depending on the yeast strains, growth media used were either medium A [0.8 g/liter CSM (Complete Supplement Mix; Bio 101)/6.5 g/liter yeast nitrogen base without amino acids (YNB) (Difco)/20 g/liter glucose] or medium B [0.8 g/liter CSM/6.5 g/liter YNB/20 g/liter galactose].
Transformation, Crossing, and Tetrad Analysis of Yeast Strains.
Transformation of S. cerevisiae was carried out essentially as described by Manivasakam and Schiestl (12). Transformants were selected on minimal selective medium [0.8 g/liter CSM lacking either tryptophan or histidine, depending on the auxotrophy selection marker of the plasmid used (Bio 101)/6.5 g/liter YNB/20 g/liter glucose]. Crossing of yeast strains of opposite mating types and tetrad analysis were done according to standard procedures (13).
Genomic DNA Library and Cloning of IPT1 Gene.
Genomic DNA from S. cerevisiae strain W303-1A was partially digested with Sau3A. Size-fractionated DNA (5–15 kb) was ligated in the yeast shuttle vector pRS423 (14) by standard methods (15). More than 14,000 clones in Escherichia coli XL2-Blue MRF′ (Stratagene) were obtained, with an average insert size of 8 kb. Plasmid DNA of pooled E. coli transformants was used to transform a DmAMP1-resistant yeast mutant DM1 (2). Twenty thousand transformants were picked up individually, transferred to microtiter plates containing yeast minimal selective medium, and grown for 48 h at 30°C. All transformants were subsequently transferred to microtiter plates containing fresh selective minimal medium including 4 μM DmAMP1 to identify DmAMP1-sensitive colonies. DmAMP1-sensitive colonies were retested in an antifungal activity assay, as described above. A plasmid preparation was made from a DmAMP1-sensitive transformant according to the method of Hoffman and Winston (16) and transformed into E. coli XL2-Blue MRF′. Plasmid DNA from the transformed E. coli cells was sequenced.
The IPT1 region of the inserted DNA fragment was subcloned in the yeast multicopy shuttle vectors pRS423 (14) and pYX233 (Ingenius R&D Systems, Madison, WI), resulting in the plasmids pRS423(IPT1) and pYX233(IPT1), respectively. For the construction of pRS423(IPT1), a 2709-bp SphI/PvuI fragment from pDM1-S encompassing the coding region of the IPT1 gene (1584 bp), a 1022-bp intergenic region containing the upstream promoter region of IPT1 and a 104-bp intergenic region containing the downstream terminator region of IPT1, was inserted into pRS423. In construct pYX233(IPT1), the inserted 1734-bp SphI/AgeI fragment from pDM1-S containing the coding region of the IPT1 gene was fused behind the galactose-inducible GAL1 promoter.
Sequencing of IPT1 Alleles in Various DmAMP1-Resistant Yeast Strains.
Genomic DNA was isolated from various DmAMP1-resistant mutant yeast strains as described by Ausubel et al. (13). The IPT1 gene of these strains was amplified by PCR with PfuTurbo polymerase (Stratagene), using primers 5′-ATGAATGTCATATTTTCTTTGGC-3′ and 5′-CTCTTATCAAACCGGCAGCAAAC-3′ for the amplification of the IPT1 coding region from 0 bp to 810 bp and primers 5′-GACACCGAACATGTTAATTACACC-3′ and 5′-CTATGCAAGCGGATCAAAAAACCA-3′ for the amplification of the IPT1 coding region from 756 bp to 1584 bp. All PCRs were done in triplicate to check for errors generated by PfuTurbo polymerase itself. The purified PCR products from each of the triplicate reactions were A-tailed with Taq DNA polymerase and ligated into the pGEM-T Easy Vector by using the pGEM-T Easy Vector Systems kit (Promega). After transformation of the ligation products in E. coli XL2-Blue MRF′, plasmids were purified and used for DNA sequencing.
SUR1 Gene Disruption.
Gene disruption of SUR1 was accomplished by replacing the coding region of SUR1 with the TRP auxotrophic marker gene as described by Brachmann et al. (17). Correct gene transplacement was verified by PCR using primers 5′-CACAGGGAGGAGGCTTACGCAGAT-3′ and 5′-TTCCAATCCAAAAGTTCACCT-3′ for the amplification of the region −245 bp upstream of the SUR1 gene to 607 bp in the TRP marker gene.
SYTOX Green Uptake and DmAMP1-Binding Assay.
Fungal membrane permeabilization was measured by SYTOX Green uptake as described previously (8). DmAMP1 was radiolabeled using t-butoxycarbonyl-l-[35S]methionine N-hydroxysuccinimidyl ester (Amersham Pharmacia) as described previously (2). Specific activities of the labeled DmAMP1 preparations were typically around 1 TBq/mmol. Binding assays with [35S]DmAMP1 on S. cerevisiae cells were performed as described previously (2).
Analysis of Sphingolipids.
The sphingolipid content of the plasma membranes of S. cerevisiae strains was performed as described previously (9). Cells were cultured overnight in medium containing 20 μCi/ml of myo-[2-3H]inositol (American Radiolabeled Chemicals, St. Louis; 20 Ci/mmol). Lipids were extracted, deacylated, and separated by TLC. Each lane on the chromatogram contained 2 μg of inositol phosphoryl ceramide (IPC)-3, IPC-4, and MIPC-3, and 1.2 μg of M(IP)2C-3 as internal standards. Radioactivity was detected by scanning the TLC plate with a Bioscan apparatus (Bioscan, Washington, DC), and the sphingolipid standards were detected by spraying the TLC plate with 10% CuSO4⋅5H2O in 8% phosphoric acid, followed by charring at 160°C.
Results
Genetic Analysis of DmAMP1-Resistant S. cerevisiae Mutants.
Previously it has been shown that growth of the yeast strain S. cerevisiae (W303-1A) is severely inhibited in the presence of the antifungal plant defensin DmAMP1 at concentrations above 1 μM (2). A spontaneous yeast mutant, called DM1, has been isolated and found to be resistant to up to 40 μM DmAMP1 (Table 1) (2). The diploid yeast strain resulting from a cross between wild type and DM1 was as sensitive to DmAMP1 as diploid wild-type yeast (Table 1), indicating that resistance conferred by the mutant allele is recessive relative to the wild-type allele. Of the 20 tetrads analyzed from wild type × DM1, all segregated as two resistant and two sensitive, which is consistent with the occurrence of a single nuclear gene conferring DmAMP1 sensitivity in wild-type yeast. An additional DmAMP1-resistant strain, DM2, had been isolated previously (2), and in the course of this work, two more resistant strains, designated as DM3 and DM4, were isolated by the same procedure. Unlike DM1, DM2, and DM3, which are fully resistant to DmAMP1 up to 40 μM, DM4 showed an intermediate level of resistance with concentrations for 50% growth inhibition (IC50) of 4.5 μM. Strains DM2, DM3, and DM4 were each crossed with DM1, and colonies grown from tetrads of the resulting diploids were tested for resistance to DmAMP1. In all cases, the resulting tetrad colonies were resistant, indicating that the mutations in strains DM2, DM3, and DM4 are allelic to that in strain DM1.
Table 1.
Antifungal activity of DmAMP1 against various S. cerevisiae strains
| S. cerevisiae strain | IC50, μM* | Medium† |
|---|---|---|
| YPH250 | 1.2 ± 0.2 | A |
| RCD113 ipt1-deletion strain | > 40 | A, B |
| W303-1A wild type (haploid) | 0.7 ± 0.1 | A |
| W303 wild type (diploid) | 1.0 ± 0.2 | A |
| DM1, DM2, and DM3 DmAMP1-resistant strains | > 40 | A |
| W303-1A × DM1 (diploid) | 1.7 ± 0.2 | A |
| DM4 DmAMP1-resistant strain | 4.5 ± 0.6 | A |
| DM1 transformed with pRS423 | > 40 | A |
| DM1 transformed with pRS423(IPT1) | 1.1 ± 0.2 | A |
| DM1 transformed with pYX233 | > 40 | B |
| DM1 transformed with pYX233(IPT1) | 1.1 ± 0.2 | B |
| RCD113 transformed with pYX233(IPT1) | 1.2 ± 0.2 | B |
| W303-1AΔsur1 | 1.4 ± 0.2 | A |
| DM1Δsur1 | 8.7 ± 0.7 | A |
IC50 values (i.e., the concentration of the antifungal protein that is required to inhibit 50% of the growth) are expressed as mean ± SE (n = 3).
Media used to grow the yeast strains were either medium A or medium B (see Materials and Methods for composition).
Taken together, these results indicate that resistance to DmAMP1 is determined by a recessive mutation in a single genetic locus, which is hereafter termed the DmAMP1 sensitivity gene.
Cloning of the DmAMP1 Sensitivity Gene.
To identify the DmAMP1 sensitivity gene, a genomic library of DmAMP1-sensitive wild-type yeast was prepared in the yeast shuttle vector pRS423 and transformed to the DmAMP1-resistant mutant yeast strain DM1. A DmAMP1-sensitive DM1 transformant was identified, and the corresponding plasmid, pDM1-S, was used to retransform DmAMP1-resistant DM1 cells. These retransformants showed the same level of DmAMP1 sensitivity as wild-type yeast, indicating that plasmid pDM1-S indeed confers sensitivity to DmAMP1. Sequence determination of the inserted DNA fragment in plasmid pDM1-S and comparison to the GenBank/EMBL nucleic acid databases revealed an insert corresponding to nucleotides 586141 to 592878 of yeast chromosome IV. This region contains three ORFs, namely, YDR070c, YDR071c, and YDR072c. ORFs YDR070c and YDR071c encode putative proteins of 93 and 191 amino acids, respectively. However, no biological function has been assigned to these ORFs. In contrast, YDR072c, an ORF of 1584 bp, encodes a plasma membrane-located protein known as IPT1. IPT1 is an enzyme that catalyzes the conversion of MIPC to M(IP)2C, the most complex and most abundant sphingolipid occurring in S. cerevisiae (9).
Sensitivity of an ipt1-Deletion Strain to DmAMP1.
To investigate a possible role of the IPT1 gene in DmAMP1 sensitivity, the ipt1-deletion strain RCD113 (9) and the corresponding wild-type strain YPH250 (10) were tested in an antifungal activity assay. The ipt1-deletion strain was resistant up to 40 μM DmAMP1, whereas wild-type strain YPH250 was sensitive to DmAMP1 at concentrations above 1 μM (Table 1). These results indicate that the DmAMP1 sensitivity gene is probably identical to IPT1.
Subcloning of IPT1.
The IPT1 gene was subcloned in the yeast shuttle vectors pRS423 and pYX233, resulting in the plasmids pRS423(IPT1) and pYX233(IPT1), respectively. As can be seen in Table 1, DM1 cells transformed with either pRS423(IPT1) or pYX233(IPT1) were sensitive to DmAMP1 at concentrations above 1 μM. In contrast, DM1 cells transformed with the insertless plasmids (either pRS423 or pYX233) were still resistant to DmAMP1 at concentrations up to 40 μM. Similarly, transformation of pYX233(IPT1) into the ipt1-deletion strain RCD113 restored sensitivity to DmAMP1 (Table 1). From these results it can be concluded that the IPT1 gene is indeed responsible for DmAMP1 sensitivity.
Sequencing of IPT1 Alleles from Various DmAMP1-Resistant Mutant Yeast Strains.
To determine the location of mutations in the IPT1 gene resulting in resistance to DmAMP1, the sequences of the IPT1 alleles of the various DmAMP1-resistant mutant yeast strains were determined and compared with the sequence of the wild-type IPT1 gene. In the sequence of the IPT1 allele from mutant DM1, the codon for Trp93 (TGG) is changed to the stop codon TAG, resulting in a truncated translation product consisting of only 92 amino acids instead of 527 amino acids. Sequence analysis of the IPT1 alleles of strains DM2, DM3, and DM4 revealed in all cases single base pair mutations leading to amino acid substitutions: Gln227 was altered to Lys in DM2; Leu268 to Phe in DM3; and Ser61 to Arg in DM4.
To make sure that DmAMP1 resistance or sensitivity is linked to the absence or presence of a functional IPT1 protein, an IPT1 gene fragment corresponding to nucleotides 173–381 of the coding region was amplified by PCR (using primers 5′-ACATTCATAGCAAGTTTGCTT-3′ and 5′-CAGATTATAGTTAACGTGCTT-3′) from 10 tetrads collected from a cross between DM1 and wild type. In each case, PCR fragments were sequenced and the resistance to DmAMP1 was determined. There was a perfect cosegregation between resistance to DmAMP1 and the presence of a stop codon in the 93rd codon of IPT1.
Binding Studies with [35S]DmAMP1.
It has been shown that binding of 35S-radiolabeled DmAMP1 to DM1 yeast cells is drastically reduced compared with binding of [35S]DmAMP1 to wild-type yeast cells (2), indicating that a reduced DmAMP1-binding capacity correlates with a reduced susceptibility to DmAMP1.
Binding of [35S]DmAMP1 was assessed on wild-type yeast cells, DM1 mutant yeast cells, and DM1 cells transformed with pRS423, pRS423(IPT1), pYX233, or pYX233(IPT1). As can be seen in Fig. 1, the [35S]DmAMP1 binding capacity of DM1 yeast cells transformed with the constructs containing the IPT1 gene was 5- to 10-fold higher compared with the [35S]DmAMP1-binding capacity of DM1 cells transformed with the control plasmids.
Figure 1.
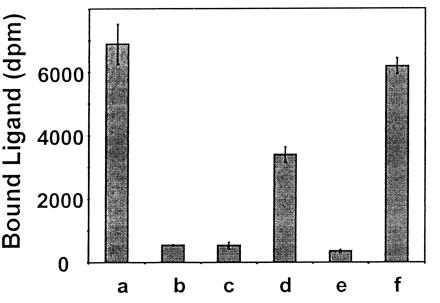
[35S]DmAMP1 binding capacity of wild-type yeast, DM1 cells, and DM1 cells transformed with pRS423, pRS423(IPT1), pYX233, and pYX233(IPT1). [35S]DmAMP1 (40 nM) was incubated with wild-type yeast cells (a), DM1 cells (b), or DM1 cells transformed with pRS423 (c), pRS423(IPT1) (d), pYX233 (e), or pYX233(IPT1) (f). Binding assays were performed after 1 h of incubation at 22°C. The growth medium was medium A for all strains, except for DM1 cells transformed with pYX233 or pYX233(IPT1), for which the galactose-containing medium B was used (see Materials and Methods for medium composition). Data are means ± SE of triplicate measurements and correspond to one representative experiment of two.
In addition, binding studies with [35S]DmAMP1 were also performed on DM2, DM3, and DM4 yeast cells. As can be seen in Fig. 2, binding of [35S]DmAMP1 to any of these DmAMP1-resistant yeast strains is drastically reduced compared with binding of [35S]DmAMP1 to wild-type yeast cells.
Figure 2.
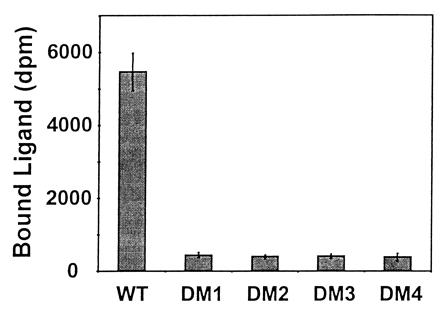
[35S]DmAMP1 binding capacity of wild-type yeast and DmAMP1-resistant yeast strains DM1, DM2, DM3, and DM4. [35S]DmAMP1 (40 nM) was incubated with wild-type yeast cells or DmAMP1-resistant yeast strains DM1, DM2, DM3, and DM4. Binding assays were performed in medium A after 1 h of incubation at 22°C. Data are means ± SE of triplicate measurements and correspond to one representative experiment of three.
Permeabilization of Wild-Type Yeast Transformed with pYX233(IPT1) and pRS423(IPT1).
To investigate the role of IPT1 in DmAMP1-mediated membrane permeabilization of yeast cells, an assay based on the uptake of SYTOX green was used as described by Thevissen et al. (8). SYTOX green is an organic compound that fluoresces upon interaction with nucleic acids and penetrates only cells with compromised plasma membranes (18, 19). By use of the SYTOX green uptake assay, it has been shown previously that DmAMP1 induces membrane permeabilization in wild-type S. cerevisiae but not in DmAMP1-resistant DM1 yeast cells (8).
DmAMP1-induced membrane permeabilization to SYTOX green was assessed for wild-type yeast, DM1 yeast cells, and DM1 cells transformed with either pYX233 or pYX233(IPT1). As can be seen in Fig. 3, SYTOX green uptake rose significantly upon treatment with DmAMP1 at concentrations above 2 μM of DM1 cells transformed with pYX233(IPT1) but not in DM1 cells or in DM1 cells transformed with the insertless vector pYX233. DmAMP1 also failed to induce permeabilization to SYTOX Green in the other DmAMP1-resistant mutants, DM2, DM3, and DM4 (results not shown).
Figure 3.
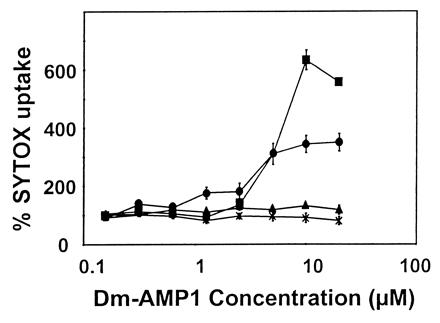
Membrane permeabilization induced by DmAMP1 on wild-type yeast, DM1 cells, and DM1 cells transformed with either pYX233 or pYX233(IPT1). Dose–response curves are presented for membrane permeabilization measured by SYTOX green fluorescence of wild-type yeast (■), DM1 cells (×), and DM1 cells transformed with either pYX233 (▴) or pYX233(IPT1) (●). Wild-type yeast and DM1 cells were suspended in medium A supplemented with 5 mM MgCl2 and 0.2 μM SYTOX green. DM1 cells transformed with either pYX233 or pYX233(IPT1) were suspended in medium B supplemented with 5 mM MgCl2 and 0.2 μM SYTOX green. Yeast cells were incubated in the presence of DmAMP1 for 12 h, after which fluorescence was measured. Data are means ± SE of triplicate measurements and correspond to one representative experiment of two.
Involvement of the Sphingolipid M(IP)2C in Resistance to DmAMP1.
The data described above indicate that the IPT1 gene determines the ability of yeast cells to bind DmAMP1 as well as to undergo DmAMP1-mediated membrane permeabilization and growth inhibition. However, the question remains whether IPT1 affects the interaction between DmAMP1 and yeast cells through the intermediary of the membrane-located IPT1 protein itself or through the membrane lipid M(IP)2C, which is synthesized by IPT1. To address this question, the capacity of wild-type yeast and the various DmAMP1-resistant yeast mutants to synthesize M(IP)2C was determined. Not surprisingly, strain DM1, which features a stop codon in the N-terminal part of the IPT1 coding region and therefore should be considered as an ipt1 null mutant, did not produce detectable amounts of M(IP)2C (Fig. 4). Furthermore, no M(IP)2C could be detected in yeast strains DM2 and DM3 (not shown), which both have a point mutation in the IPT1 coding region and are resistant to up to 40 μM DmAMP1. In the case of yeast mutant DM4, which is resistant to up to 4 μM DmAMP1, a small amount of M(IP)2C was detected (Fig. 4). Hence partial resistance to DmAMP1 correlates with a low level of M(IP)2C, whereas full resistance correlates with absence of M(IP)2C. To further discriminate between the IPT1 protein and M(IP)2C as possible determinants of sensitivity to DmAMP1, sur1-deletion strains of wild-type yeast and of the ipt1 null mutant DM1 were constructed. SUR1 is a mannosyltransferase that catalyzes the penultimate step in M(IP)2C biosynthesis, more precisely, the conversion of IPC to MIPC (20). It was shown previously that deletion of the SUR1 gene prevents synthesis of the mannosylated sphingolipids M(IP)2C and MIPC and instead causes the sphingolipid IPC to accumulate in the membranes (20). The sur1-deletion mutant of DM1, DM1Δsur1, was sensitive to DmAMP1 at concentrations above 9 μM, whereas DM1 was resistant to up to 40 μM (Table 1). Because the two strains lack IPT1 but differ in their sphingolipid composition (DM1 contains mainly MIPC as sphingolipid, whereas DM1Δsur1 contains only IPC), the difference in sensitivity to DmAMP1 must be due at least in part to a difference in sphingolipid composition. The difference in the strains also implies that IPC can at least partially substitute for M(IP)2C as a determinant for DmAMP1 sensitivity. This implication is also in line with the observation that the sur1-deletion strain of wild-type yeast, W303-1AΔsur1, is only 2-fold less sensitive to DmAMP1 than is wild-type yeast (Table 1).
Figure 4.
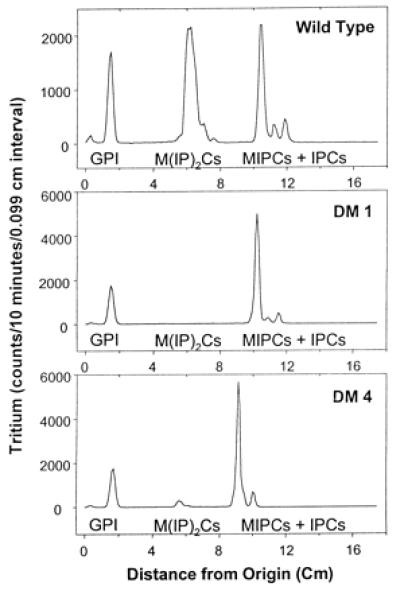
DmAMP1-resistant yeast strains lack the sphingolipid M(IP)2C. Preparation of [3H]inositol-labeled cells, lipid extraction, TLC, tritium detection, and location of internal sphingolipid standards are described in Materials and Methods. The location of internal standards is indicated below the peaks, as is the location of glycerol phosphoinositol (GPI). Strains DM2 and DM3 gave a TLC profile identical to that of strain DM1 (data not shown). Differences in migration distances are due to lane-to-lane variation in mobility.
Discussion
This paper describes the isolation and characterization of a gene from S. cerevisiae that determines the sensitivity of this yeast to the antifungal plant defensin DmAMP1. This gene was identified by screening for restored sensitivity to DmAMP1 of a DmAMP1-resitant yeast mutant complemented with a library of genomic DNA fragments prepared from a wild-type DmAMP1-sensitive yeast strain. Several lines of evidence show unequivocally that the gene determining the sensitivity of S. cerevisiae to DmAMP1 is IPT1, a gene involved in sphingolipid biosynthesis: (i) yeast strains with null alleles of IPT1, as the result of a deletion or a nonsense point mutation in this gene, are fully resistant to DmAMP1; (ii) a null allele of IPT1 cosegregates with resistance to DmAMP1 in the progeny of a cross between an ipt1 null mutant and wild-type yeast; (iii) strains with null alleles of IPT1 become sensitive to DmAMP1 when the wild-type IPT1 allele is reintroduced on a plasmid. Furthermore, we have shown that strains with an ipt1 null allele show a reduced binding capacity for radiolabeled DmAMP1 and are resistant to DmAMP1-mediated permeabilization of their membranes. Both binding capacity for DmAMP1 and permeabilization upon treatment with DmAMP1 are restored when the ipt1 null mutants are complemented with a wild-type IPT1 allele.
Our finding that IPT1 determines DmAMP1-binding capacity, DmAMP1-mediated permeabilization, as well as DmAMP1-mediated growth inhibition provides strong support for a model in which all three phenomena are causally linked. In such a model, DmAMP1 would interact with a binding site on the plasma membrane, which would facilitate insertion in the plasma membrane, with subsequent formation of structures that alter membrane permeability. We have previously shown that specific binding sites for DmAMP1 reside on the plasma membrane of fungi and that such binding cannot be reversed once binding has taken place (2), which is consistent with the presumed membrane insertion event. We have also demonstrated that treatment of fungi with DmAMP1 alters the permeability of their membranes nonselectively to allow passage of ions such as Ca2+ and K+ (7) or small solutes such as the fluorescent dye SYTOX green (8) and, furthermore, that the dose–response curves of DmAMP1-mediated permeabilization correlate tightly with those for DmAMP1-mediated growth inhibition (8).
IPT1 is a biosynthetic enzyme that catalyzes the conversion of the membrane sphingolipid MIPC into M(IP)2C. Sphingolipids are, along with sterols and phosphoglycerolipids, one of the three major types of lipids found in membranes of eukaryotes. In the yeast S. cerevisiae, three types of sphingolipids are synthesized: IPC, MIPC, and M(IP)2C. These three types of sphingolipids are located primarily in the plasma membrane (21, 22), where they account for 30% of the plasma membrane phospholipids. M(IP)2C, the terminal sphingolipid, represents the major fraction among membrane sphingolipids (21). Sphingolipids associate with sterols in the plasma membrane to form patches (also called rafts) that are highly enriched in glycosyl-phosphatidylinositol (GPI)-anchored membrane proteins (23). Processing and correct targeting of these proteins during transport from the endoplasmic reticulum to the plasma membrane have been shown to require sphingolipids (23, 24). Other presumed roles of sphingolipids include signal transduction during the heat stress response and regulation of calcium homeostasis or components in calcium-mediated signaling pathways (25).
A possible role for the IPT1 protein in DmAMP1 susceptibility is its involvement in constituting the DmAMP1 binding site on the fungal plasma membrane. However, the cellular location of the IPT1 protein has not been determined, but it is most likely in the Golgi apparatus (9) and is therefore not a likely candidate for being the DmAMP1 binding site on the yeast plasma membrane. Furthermore, three different DmAMP1-resistant yeast mutants (DM2, DM3, and DM4) have IPT1 allozymes with amino acid substitutions at positions 227, 268, and 61, respectively, leading in all three cases to reduced or abolished M(IP)2C production. In addition, an independently identified yeast mutant lacking M(IP)2C, mic2 (26), was found by us to be resistant to DmAMP1 and to have an amino acid substitution at position 272 of the IPT1 enzyme (results not shown). Hence, all of these mutations affect resistance to DmAMP1 and the catalytic activity of the enzyme. In hypothesizing that DmAMP1 interacts directly with IPT1 one should assume that residues 61, 227, 268, and 272 are also involved in binding to DmAMP1, which would be a very unlikely coincidence. Moreover, residue 61 is located in a predicted transmembrane domain of the enzyme (27), a location that is consistent with its involvement in binding or conversion of the sphingolipid substrate but not with a role in protein–protein interactions.
A DM1 yeast strain derivative with a deleted SUR1 gene, encoding the enzyme that catalyzes the conversion of IPC into MIPC, was found to be sensitive to DmAMP1 at concentrations above 9 μM. Membranes of this DM1sur1-deletion strain contain the sphingolipid IPC but lack MIPC and M(IP)2C, whereas DM1 contains mainly MIPC. Because both DM1 and DM1Δsur1 do not have a functional IPT1 protein, it can be concluded that resistance to DmAMP1 has to do with modifications in sphingolipid composition rather than with alterations of the IPT1 structure. Strains with M(IP)2C (wild-type yeast strains) are highly sensitive to DmAMP1, strains with mainly MIPC (ipt1-deletion mutants) are highly resistant, and those with mainly IPC (sur1-deletion mutants) are sensitive, although slightly less so than wild-type strains (Table 1). We conclude from this observation that membrane patches with either M(IP)2C or IPC are determinants for DmAMP1 sensitivity. However, we do not exclude the possibility that IPT1 catalyzes the synthesis of inositol-containing membrane lipids other than M(IP)2C. Indeed, our observation that the ipt1, sur1 double mutant is about 6 times more resistant to DmAMP1 than the sur1 mutant calls for a role of IPT1 in the conversion of substrates other than MIPC into molecules that play a role in DmAMP1 sensitivity.
The sphingolipids IPC and/or M(IP)2C might interact directly with DmAMP1 and facilitate its insertion into the membrane. The terminal inositol group in IPC and M(IP)2C, masked by the presence of mannose in the case of MIPC, might be important for establishing this interaction. Sphingolipids have not been identified so far as possible targets for antimicrobial peptides. Most antimicrobial peptides studied thus far are known to interact with ubiquitous phosphoglycerolipids (28–32), explaining their relatively broad antimicrobial spectrum. Nisin, a narrow-spectrum antibacterial peptide produced by some lactic acid bacteria, has been shown to interact specifically with the membrane-bound peptidoglycan precursor lipid II. Lipid II acts as a docking site for nisin, facilitating its subsequent insertion into the bacterial membrane (33).
An alternative hypothesis that is equally well supported by our data is that IPC and/or M(IP)2C is required for stabilizing or exposing particular GPI-anchored membrane proteins in the plasma membrane. Such GPI-anchored proteins would then act as docking sites for DmAMP1 and assist in membrane insertion of DmAMP1. The inositol group of IPC and M(IP)2C may be important for the presumed interaction with the GPI-anchored membrane proteins. GPI-anchored proteins on mammalian cells have previously been shown to act as high-affinity receptors for aerolysin, one of the major toxins secreted by the bacterium Aeromonas hydrophila. Aerolysin interacts with the GPI anchor of GPI-anchored proteins, resulting in multimerization of the toxin and subsequent pore formation (23).
The possible involvement of GPI-anchored proteins in the DmAMP1 sensitivity of yeast can be studied by comparing membrane and cell-wall protein profiles of ipt1 null mutants and wild-type yeast. In addition, it should be informative to investigate the possibility that DmAMP1 interacts directly with the sphingolipids M(IP)2C and IPC, by using artificial liposomes supplemented with purified sphingolipids.
Acknowledgments
We thank Dr. G. Daum for the mic2 yeast mutant. This research was supported in part by the Fonds voor Wetenschappelijk Onderzoek–Vlaanderen (Grant G.0218.97) and by a grant (GM 41302) from the National Institutes of Health (Bethesda, MD) (to R.C.D.). K.T. acknowledges receipt of a Postdoctoral Research Fellowship from the Onderzoeksfonds of the Katholieke Universiteit of Leuven, Belgium.
Abbreviations
- AMP
antimicrobial protein/peptide
- DmAMP1
Dahlia merckii AMP 1
- IPC
inositol phosphorylceramide
- MIPC
mannose-(inositol-phosphate)-ceramide
- M(IP)2C
mannose-(inositol-phosphate)2-ceramide
- GPI
glycosyl-phosphatidylinositol
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.160077797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.160077797
References
- 1.Boman H G. Scand J Immunol. 1998;48:15–25. doi: 10.1046/j.1365-3083.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 2.Thevissen K, Osborn R W, Acland D P, Broekaert W F. Mol Plant–Microbe Interact. 2000;13:54–61. doi: 10.1094/MPMI.2000.13.1.54. [DOI] [PubMed] [Google Scholar]
- 3.Broekaert W F, Terras F R, Cammue B P, Osborn R W. Plant Physiol. 1995;108:1353–1358. doi: 10.1104/pp.108.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broekaert W F, Cammue B P A, De Bolle M F C, Thevissen K, De Samblanx G W, Osborn R W. Crit Rev Plant Sci. 1997;16:297–323. [Google Scholar]
- 5.Harrison S J, Marcus J P, Goulter K C, Green J L, Maclean D J, Manners J M. Aust J Plant Physiol. 1997;24:571–578. [Google Scholar]
- 6.Osborn R W, De Samblanx G W, Thevissen K, Goderis I, Torrekens S, Van Leuven F, Attenborough S, Rees S B, Broekaert W F. FEBS Lett. 1995;368:257–262. doi: 10.1016/0014-5793(95)00666-w. [DOI] [PubMed] [Google Scholar]
- 7.Thevissen K, Ghazi A, De Samblanx G W, Brownlee C, Osborn R W, Broekaert W F. J Biol Chem. 1996;271:15018–15025. doi: 10.1074/jbc.271.25.15018. [DOI] [PubMed] [Google Scholar]
- 8.Thevissen K, Terras F R, Broekaert W F. Appl Environ Microbiol. 1999;65:5451–5458. doi: 10.1128/aem.65.12.5451-5458.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickson R C, Nagiec E E, Wells G B, Nagiec M M, Lester R L. J Biol Chem. 1997;272:29620–29625. doi: 10.1074/jbc.272.47.29620. [DOI] [PubMed] [Google Scholar]
- 10.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broekaert W F, Terras F R G, Cammue B P A, Vanderleyden J. FEMS Microbiol Lett. 1990;69:55–60. [Google Scholar]
- 12.Manivasakam P, Schiestl R H. Nucleic Acids Res. 1993;21:4414–4415. doi: 10.1093/nar/21.18.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1993. [Google Scholar]
- 14.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 15.Philippsen P, Stotz A, Scherf C. Methods Enzymol. 1991;194:169–182. doi: 10.1016/0076-6879(91)94014-4. [DOI] [PubMed] [Google Scholar]
- 16.Hoffman C S, Winston F. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 17.Brachmann C B, Davies A, Cost G J, Caputo E, Li J, Hieter P, Boeke J D. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 18.Matsuzaki T, Suzuki T, Fujikura K, Takata K. Acta Histochem Cytochem. 1997;30:309–314. [Google Scholar]
- 19.Roth B, Poot M, Yue S, Millard P. Appl Environ Microbiol. 1997;63:2421–2431. doi: 10.1128/aem.63.6.2421-2431.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beeler T J, Rivera D F J, Monaghan E, Gable K, Dunn T M. Mol Gen Genet. 1997;255:570–579. doi: 10.1007/s004380050530. [DOI] [PubMed] [Google Scholar]
- 21.Patton J L, Lester R L. J Bacteriol. 1991;173:3101–3108. doi: 10.1128/jb.173.10.3101-3108.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hechtberger P, Zinser E, Saf R, Hummel K, Paltauf F, Daum G. Eur J Biochem. 1994;225:641–649. doi: 10.1111/j.1432-1033.1994.00641.x. [DOI] [PubMed] [Google Scholar]
- 23.Bagnat M, Keranen S, Shevchenko A, Shevchenko A, Simons K. Proc Natl Acad Sci USA. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skrzypek M, Lester R L, Dickson R C. J Bacteriol. 1997;179:1513–1520. doi: 10.1128/jb.179.5.1513-1520.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dickson R C. Annu Rev Biochem. 1998;67:27–48. doi: 10.1146/annurev.biochem.67.1.27. [DOI] [PubMed] [Google Scholar]
- 26.Leber A, Fischer P, Schneiter R, Kohlwein S D, Daum G. FEBS Lett. 1997;411:211–214. doi: 10.1016/s0014-5793(97)00692-3. [DOI] [PubMed] [Google Scholar]
- 27.Dickson R C, Lester R L. Biochim Biophys Acta. 1999;1438:305–321. doi: 10.1016/s1388-1981(99)00068-2. [DOI] [PubMed] [Google Scholar]
- 28.Cociancich S, Ghazi A, Hetru C, Hoffmann J A, Letellier L. J Biol Chem. 1993;268:19239–19245. [PubMed] [Google Scholar]
- 29.Kagan B L, Selsted M E, Ganz T, Lehrer R I. Proc Natl Acad Sci USA. 1990;87:210–214. doi: 10.1073/pnas.87.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ladokhin A S, Selsted M E, White S H. Biophys J. 1997;72:794–805. doi: 10.1016/s0006-3495(97)78713-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuzaki K, Sugishita K, Harada M, Fujii N, Miyajima K. Biochim Biophys Acta. 1997;1327:119–130. doi: 10.1016/s0005-2736(97)00051-5. [DOI] [PubMed] [Google Scholar]
- 32.Tejuca M, Serra M D, Ferreras M, Lanio M E, Menestrina G. Biochemistry. 1996;35:14947–14957. doi: 10.1021/bi960787z. [DOI] [PubMed] [Google Scholar]
- 33.Brotz H, Josten M, Wiedemann I, Schneider U, Gotz F, Bierbaum G, Sahl H G. Mol Microbiol. 1998;30:317–327. doi: 10.1046/j.1365-2958.1998.01065.x. [DOI] [PubMed] [Google Scholar]


