Abstract
Scansite identifies short protein sequence motifs that are recognized by modular signaling domains, phosphorylated by protein Ser/Thr- or Tyr-kinases or mediate specific interactions with protein or phospholipid ligands. Each sequence motif is represented as a position-specific scoring matrix (PSSM) based on results from oriented peptide library and phage display experiments. Predicted domain-motif interactions from Scansite can be sequentially combined, allowing segments of biological pathways to be constructed in silico. The current release of Scansite, version 2.0, includes 62 motifs characterizing the binding and/or substrate specificities of many families of Ser/Thr- or Tyr-kinases, SH2, SH3, PDZ, 14-3-3 and PTB domains, together with signature motifs for PtdIns(3,4,5)P3-specific PH domains. Scansite 2.0 contains significant improvements to its original interface, including a number of new generalized user features and significantly enhanced performance. Searches of all SWISS-PROT, TrEMBL, Genpept and Ensembl protein database entries are now possible with run times reduced by ∼60% when compared with Scansite version 1.0. Scansite 2.0 allows restricted searching of species-specific proteins, as well as isoelectric point and molecular weight sorting to facilitate comparison of predictions with results from two-dimensional gel electrophoresis experiments. Support for user-defined motifs has been increased, allowing easier input of user-defined matrices and permitting user-defined motifs to be combined with pre-compiled Scansite motifs for dual motif searching. In addition, a new series of Sequence Match programs for non-quantitative user-defined motifs has been implemented. Scansite is available via the World Wide Web at http://scansite.mit.edu.
INTRODUCTION
Characterizing protein interactions on a proteome-wide scale is required to catalyze the advance of systems biology. Online databases of protein sequences (1–5) and known protein–protein interactions (6–8) are the first steps taken in this direction, but finding new interactions will require new combinations of experimental and computational methods. Scansite (http://scansite.mit.edu) is a computational tool built on experimental binding and/or substrate information from oriented peptide library screening (9–13) and phage display experiments (14), together with detailed biochemical characterization to derive a weight matrix-based scoring algorithm that predicts protein–protein interactions and sites of phosphorylation (15).
DOMAINS AND MOTIFS
The accumulated molecular structures in the Protein Data Bank (PDB) make it clear that eukaryotic proteins are often built with a modular architecture, combining domains that fold and function independently into larger polypeptides. These domains often occur in multiple unrelated proteins, where they fulfill similar targeting functions. Identification of these domains within a protein can be a valuable indicator of the function of the protein as a whole and can assist in placing that protein within the correct cell signaling pathway. A number of modular domains such as WW, SH2, SH3, PTB, PDZ and 14-3-3 bind to their ligands through direct interactions with very short amino acid sequences (typically <10 amino acids), or in the case of protein kinases, phosphorylate a Ser-, Thr- or Tyr-containing sequence motif in their protein substrates. Modular binding domains are typically fairly long (60–300 residues) and can be identified using sequence comparison methods and Hidden Markov Models [e.g. Pfam (16) and SMART (17)]. In contrast, the corresponding motifs to which they bind are much shorter (3–10 residues) and have been more elusive to locate. The current release (version 7.8) of Pfam, for example, identifies 4941 protein domains and families, but only 18 motifs (16). Scansite was developed to address this need and to facilitate work in our own laboratories on signaling by protein kinases and modular phosphopeptide- and phospholipid-binding domains.
Many of the motifs in Scansite were determined using oriented peptide library experiments. In this technique, degenerate peptides with a single fixed (orienting) central residue are incubated with one type of domain (9–13). Because of our laboratories' research focus, this central residue was typically a Ser, Thr or Tyr for protein kinase domains, or a phosphoSer/Thr or phosphoTyr residue for phosphospecific binding domains (such as SH2, PTB or 14-3-3 domains). Peptides that were phosphorylated by the kinase or were bound by the binding domain were isolated and sequenced as an ensemble by Edman degradation. When sequenced in this manner, each Edman cycle reveals the relative amount of each amino acid residue occurring at that position. This information is then scaled and normalized to produce a scoring matrix (i.e. a PSSM) which quantitatively indicates the preference for each amino acid type at each position within the domain's recognition motif. These matrices can then be used to score entire databases of protein sequences to find a small number of proteins with high-ranking motif matches, indicating possible protein–protein interactions. As the number of motifs grew, the opposite search became practical as well: scanning a single protein sequence for matches to any of the motifs in our database.
We have collected these programs to create a user-friendly web-based tool accessible to the entire scientific community that allows investigators to search for motifs recognized by commonly occurring domains within a protein sequence query of their choice or to search entire protein sequence databases for optimal motif matches. The Motif Scan ensemble of programs computationally identifies all motifs within a given user-specified protein, while the Database Search ensemble of programs finds all proteins in a protein database, such as SWISS-PROT, that match a given motif. By repeated queries using the results of one search to launch another, it is possible to infer several steps of a signaling pathway in silico. For example, if a newly discovered protein is predicted by Scansite to be phosphorylated by the kinase domain from Akt and the resulting phosphorylation is predicted to create a binding site for 14-3-3 proteins, then the newly discovered protein is likely to function in a signaling pathway involving these proteins. These types of analyses performed on protein sequence databases can functionally annotate a limited number of promising interactions that merit experimental investigation and may also suggest that other intermolecular interactions are unlikely, at least within the limits of sequence-based prediction.
STRINGENCY LEVELS
Threshold values need to be assigned when scanning query proteins with the Motif Scan programs to decide which scores are likely to suggest real interactions. Scansite incorporates three settings, labeled ‘high’, ‘medium’ and ‘low’ stringencies; the high stringency setting is the most restrictive and reports a ‘hit’ only if the score falls within the top 0.2% of scores when the motif matrix of interest was applied to the vertebrate subset of SWISS-PROT. This dataset was chosen as a reference because of the non-redundant nature of SWISS-PROT and the relevance of vertebrate proteins to the type of cell signaling events predicted by Scansite. These values were found to increase the reliability of prediction of true positive ‘hits’ while minimizing the number of predicted false negative interactions, based on a comparative analysis of mammalian and bacterial database subsets (15). The medium and low stringencies were then arbitrarily chosen at 1 and 5%, respectively.
Scoring percentiles in the Database Search programs, on the other hand, are calculated de novo, based solely on the protein database subset selected for the search. For example, a search among human proteins will yield sites whose percentiles are relative to all human proteins included in the search. The same site can thus have a different percentile for different database searches, but its score is always constant.
It should always be borne in mind by the user that Scansite predicitions are based solely on 1D sequence comparison and all predicted interactions must be experimentally verified before they can be considered valid.
MATERIALS AND METHODS
Server
The public collection of Scansite programs runs on a Dell PowerEdge 8450 server, with 8 Intel Xeon 733 MHz CPUs and 4 Gb of RAM. Two 32 Gb hard drives are used in a RAID 1 array. The operating system is Red Hat Linux 7.3.
Development
All development for Scansite version 2.0 was performed using the GNU GCC compiler, the PHP 4.0 and Perl 5.5 interpreters, Mandrake Linux 8.0 through 9.0, Red Hat Linux 7.1 through 7.3, the Apache 1.3 web server, the MySQL 3.23 relational database and the KDE desktop environment.
SCANSITE PROGRAMS
A total of 10 programs are included in Scansite 2.0 and these are listed in Table 1. The Motif Scan programs can accept either a protein accession number or a sequence as input and can optionally accept a user-defined motif. The Database Search programs can operate on one or more Scansite motifs, one or more user-defined motifs or combinations of Scansite and user-defined motifs. The Quick Matrix Method allows users to construct a roughly quantitative matrix based on qualitative residue preferences for a sequence motif. The Sequence Match programs allow users to find occurrences of one or two specified consensus sequences in the protein databases and can also be used to find any MySQL-recognized regular expression. A brief description of using each of these programs follows. More detailed instructions can be found in the tutorial on our web site (http://scansite.mit.edu/tutorial/tutorial.html) (see also 18).
Table 1. List of programs available on the Scansite web site.
| Motif Scan | Scans a protein sequence for motifs |
| Scan a Protein by Accession Number or ID | Takes accession number or ID as input (e.g. RB_HUMAN, P06400) |
| Scan a Protein by Input Sequence | Takes sequence as input |
| Scan Input Sequence with an Input Motif | Takes sequence and a user-defined motif as input |
| Database Search | Searches a database for motifs |
| Search Using a Scansite Motif | Searches for a single pre-compiled Scansite motif |
| Search Using an Input Motif | Takes a user-defined motif as input |
| Search Using Quick Matrix Method for Making a Motif | Takes a semi-quantitative user-defined motif as input |
| Search Using Multiple Motifs | Searches for multiple pre-compiled or user-defined motifs |
| Sequence Match | Retrieves all sequences matching an input pattern exactly |
| Search Databases for Sequence Pattern | Takes a single sequence pattern as input |
| Search Databases for Two Sequence Patterns | Takes two sequence patterns as input |
| Search Databases for Regular Expression | Takes a regular expression as input |
Motif Scan
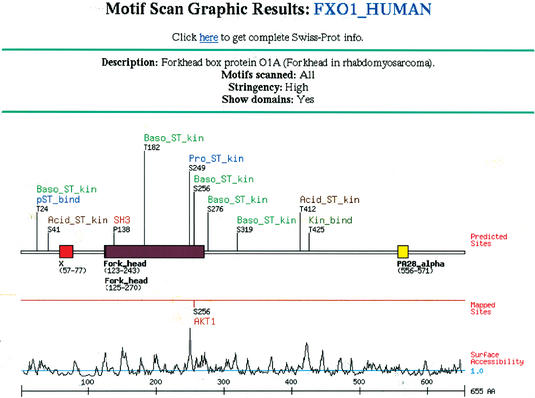
To use the Motif Scan programs, users should go to the web site http://scansite.mit.edu. Under the heading ‘Motif Scan’, click ‘Scan a Protein by Accession Number or ID’ to use a protein from a public database or click ‘Scan a Protein by Input Sequence’ to enter a protein sequence directly. The required inputs are then displayed, which include the protein's accession number and database of origin (or with the input sequence version, the protein's sequence and an arbitrary name for it), followed by the list of motifs to scan for. The default setting is to search for occurrences of all motifs in the Scansite database. Alternatively, one or more individual motifs can be selected, or several motifs of similar type (i.e. a ‘motif group’) can be selected at once. The list of motifs currently available in Scansite is shown in Table 2. Users can search at high stringency (the default choice), which shows only the strongest motif matches or at medium or low stringency to see weaker sites. Finally, users can elect to identify domains in the protein sequence, which Scansite accomplishes by parsing the results from an external call to the Pfam server at Washington University, St Louis (16). This lengthens the time needed to generate results, but the domain information is often very informative. With all these settings selected, clicking the ‘Submit Request’ button initiates the scan. The result will show a schematic map of the protein with the predicted sites found (Fig. 1) and a detailed table showing the score and sequence of each one (Fig. 2).
Table 2. Current list of motifs included in Scansite.
| Phosphoserine/threonine binding domains | 14-3-3 mode 1 |
| Tyrosine kinase domains | Abl |
| EGFR | |
| FGFR | |
| Insulin receptor | |
| Itk | |
| Lck | |
| PDGFR | |
| Src | |
| Src homology 2 domains | Abl |
| Crk | |
| FGFR | |
| Fyn | |
| Grb2 | |
| Itk | |
| Lck | |
| Nck | |
| p85 | |
| PLC γ (C terminal SH2) | |
| PLC γ (N terminal SH2) | |
| Shc | |
| SHIP | |
| Src | |
| Src homology 3 domains | Abl |
| Amphiphysin | |
| Cbl-associated protein | |
| Cortactin | |
| Crk | |
| Grb2 | |
| Intersectin | |
| Itk | |
| Nck | |
| p85 α (mode 1) | |
| p85 α (mode 2) | |
| PLC γ | |
| Src | |
| Basophilic serine/threonine kinase domains | Akt |
| Calmodulin-dependent kinase 2 | |
| Clk2 | |
| Protein kinase A | |
| PKC α/β/γ | |
| PKC δ | |
| PKC ɛ | |
| PKC μ | |
| PKC ζ | |
| DNA damage kinase domains | ATM |
| DNA protein kinase | |
| Acidophilic serine/threonine kinase domains | Casein kinase 1 |
| Casein kinase 2 | |
| GSK3 | |
| Proline-dependent serine/threonine kinase domains | Cdc2 |
| Cdk5 | |
| Erk1 | |
| p38 MAP kinase | |
| Kinase binding domains | Erk1 |
| PDK1 | |
| PDZ binding domains | PDZ class 1 |
| PDZ class 2 | |
| PDZ (nNOS) class 1 | |
| PDZ (nNOS) class 3 | |
| Phosphotyrosine binding domains | Shc |
| Lipid binding domains | PIP3-binding PH |
Figure 1.
Description of elements in Motif Scan graphical output. The protein query (in this case, the transcription factor FOXO1) is represented schematically as a line, with colored rectangles marking known domains. Labels above the protein indicate where motifs were found and identify the motif family. Labels below the protein indicate the name and range of each domain found. If the protein's annotation includes phosphorylation sites that have been experimentally mapped (generally true only for some SWISS-PROT entries), these are also indicated below the domains. On the next line, a plot of the predicted surface accessibility at each residue, calculated using a 6 amino acid running window (19) is shown. The ruler at the bottom marks numbered intervals along the protein sequence.
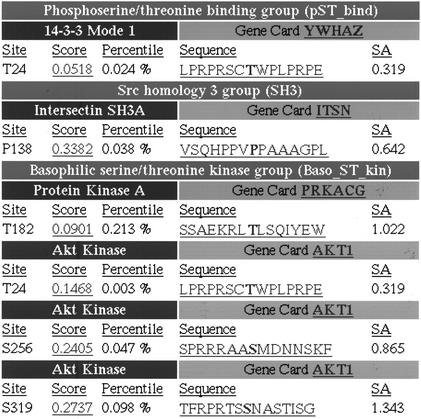
Figure 2.
Motif Scan's output table. For each motif family with a site on the graphical output (cf Fig. 1), details about the best matching domain motifs and the position of the site in the query are shown. The score, percentile and sequence of the site are indicated, as is the calculated surface accessibility for that site (labeled SA). Clicking on the score will display a histogram showing where this score ranks when compared with all potential sites for that motif in vertebrate SWISS-PROT; clicking on the sequence shows its position in the full protein and provides a link to BLAST for evaluating conservation of the motif in related protein homologues. For domains with an entry in the Weizman Institute's GeneCard database (http://bioinformatics.weizmann.ac.il/cards), the name is listed as a hyperlink to its GeneCard reference.
Database Search
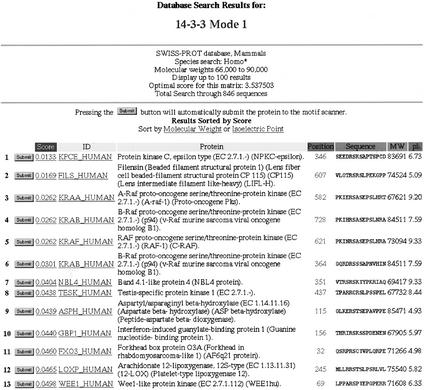
To use the Database Search program, users should click ‘Search Using a Scansite Motif’ under the ‘Database Search’ heading. A list of all the motifs in Scansite is shown. Users should select one of the motifs to search with and select the name of the protein database to search. The databases currently available are SWISS-PROT, TrEMBL, Genpept and Ensembl. Optionally, the search can be limited to proteins in just one species or a category of organisms, including mammals, vertebrates, invertebrates, plants, fungi, viruses and bacteria and archaea (grouped together). Other options allow searching within a specified range of molecular weights and isoelectric points, to facilitate comparison with two-dimensional gel electrophoresis experiments. Restricting the results by keywords in the protein description and/or by characteristic subsequences is also possible. The last user-specified parameter is the desired size of the search output, ranging from 50 to 2000 reported sites. Clicking ‘Submit Request’ starts the search. The resulting table (Fig. 3) lists all sites found, identifying the associated protein's name, description, sequence, molecular weight and isoelectric point. Any protein found from a database search can be rapidly submitted to the Motif Scan program by clicking the ‘Submit’ button on the far left of each output line.
Figure 3.
Output table from a Database Search. The name of the motif used in the search (in this case 14-3-3) is displayed at the top, with any search restictions specified immediately below (in this case, human proteins with Mw from 66 to 90 kDa). Each line in the table lists the score and sequence of a site found, together with its protein ID, description, molecular weight and isoelectric point. Clicking the Submit button at the left launches the Motif Scan program for that protein to facilitate further analysis. This table is sorted by score, but can alternatively be sorted by molecular weight or isoelectric point.
In addition to the pre-compiled Scansite motifs listed, investigators can use their own motifs to search databases, using the program ‘Search Using an Input Motif’. A tab-delimited text file containing a weight matrix is uploaded into Scansite and the subsequent options and output are the same as described above. Instructions on how to create and upload a matrix are provided in the tutorial page on our web site (http://scansite.mit.edu/tutorials/tutorial.html).
One variation on the Database Search is the program ‘Search Using Quick Matrix Method’. This program allows users to define an approximate motif by specifying a short pattern of amino acids, where wildcards are allowed. For a motif such as RXSXL, this sequence can be entered in the row of positions labeled ‘Primary Preference’. Optionally, if it was known that proline can substitute for the leucine, a P can be entered in the ‘Secondary Preference’ row at that position. Scansite makes a crude weight matrix based on these inputs, assigning a score of 9.0 to residues in the primary preference row, a score of 4.5 to those in the secondary preference row and a score of 1.0 to all unspecified residues. The results of using the Quick Matrix Method will be less quantitative than a normal database search, but can yield useful results when only limited motif information is available.
Sequence Match
The Sequence Match programs are new in the current release of Scansite. As with the Quick Matrix Method, these programs are useful when only partial motif information is available. Unlike the Quick Matrix Method, these programs do not provide quantitative match ranking, but they instead retrieve all proteins in a database that exactly match the sequence pattern specified, similar to the programs Patscan (Ross Overbeek and Alex Rodriguez, http://www-unix.mcs.anl.gov/compbio/PatScan/HTML/patscan.html) and ScanProsite (http://us.expasy.org/tools/scanprosite/). Unlike those two programs, Sequence Match will accept the widely used regular expression syntax common in Perl, PHP, MySQL and other programming environments. This kind of information can help an investigator decide how rare or specific a hypothetical motif is, how functionally similar the proteins containing the motif are, whether a motif occurs more commonly in one species or another and how many proteins may cross-react with an antibody made using the motif as an epitope. As with the Database Search, the proteins retrieved can be limited to the most relevant ones by specifying a single species, molecular weight range and values for the other options mentioned previously.
There are three Sequence Match programs. The first and simplest takes a single consensus sequence as input, which may contain wildcards. The second program looks for two different consensus sequences occurring simultaneously in the same protein. The third and most flexible program is ‘Search Databases for Regular Expression’. Unlike the first two programs, this program allows gaps of any length, alternative residues at any position and motifs at the N- or C-termini of proteins (such as signal sequences or antibody epitopes). Any regular expression recognized by MySQL can be used as the search term and our web site gives the full list of allowed symbols as well as several biologically useful examples.
IMPROVEMENTS IN VERSION 2.0 OF SCANSITE
Speed
Program execution speed has been significantly improved for the Database Search programs. Storing protein sequence information in a relational database rather than in text files, in combination with rewriting the base code, shortened the time needed for a typical database search by approximately a factor of three compared with Scansite version 1.0. Our protein sequence databases are currently updated with each major release of Genpept, SWISS-PROT, TrEMBL and Ensembl. Between updates, very recent additions to these databases may not be present in Scansite.
Targeted searches
In addition to speed, the MySQL relational databases for protein sequences and motif PSSMs facilitate restricted database searches based on pre-annotated database entries. Scansite 2.0 gives researchers the ability to find motifs in proteins from a single species or genus, within a range of molecular weights and isoelectric points, or containing keywords, and/or a characteristic subsequence (which can lie outside the motif region). The Motif Scan programs similarly benefit: rather than searching for all motifs or individually selected ones, users can now search by motif ‘groups’, where functionally similar motifs have been grouped together (e.g. SH2 domains, SH3 domains, tyrosine kinases and others) (Table 2). One or more motif groups can also be combined with one or more individually selected motifs.
Graphics
The algorithm previously used to display sites and domains graphically along the protein sequence sometimes led to overlapping text, making annotations difficult to read. The new algorithm displays many more sites and domains without overlap. In response to numerous user requests, the generated graphic is now a single downloadable PNG image to facilitate publication of users' results.
Two-dimensional gel electrophoresis
Results from a Database Search can be sorted by molecular weight or isoelectric point and the search can be restricted to proteins within a narrow range of both parameters. As a result, Scansite can be used in conjunction with two-dimensional gel electrophoresis experiments to help identify spots in regions of a gel. For experiments involving primarily phosphoproteins, the expected number of phosphate groups can be specified in the Database Search options and mass and isoelectric point calculations correspondingly adjusted.
User-entered motifs
Users have always been able to enter their own motifs to perform Scansite searches. In version 2.0, we made three additions. First, we now allow use of matrices that lack values for one or more amino acid types by supplying default values for those positions. Second, researchers studying selenocysteine-containing proteins can now enter motifs giving a score for selenocysteine by labeling that column ‘U’, its accepted single-letter code. Third, motifs targeting the N-terminus of a protein sequence can now be specified, using a column labeled with the arbitrarily chosen character ‘$’ (dollar sign). The ability to use C-terminal-directed motifs has existed since version 1.0 by using the ‘*’ character and is currently used in PSSMs for PDZ domains.
Multiple motifs
Searching for proteins that contain motifs of more than one type can be a powerful way to increase the functional relevance of database searches (15). Version 1.0 allowed users to search for two Scansite motifs or two user-entered motifs. Version 2.0 allows users to search for proteins containing up to five different motifs, which can be any combination of Scansite motifs and user-entered motifs.
User-contributed motifs
The Database Search, Quick Matrix Method and Sequence Match programs allow users to temporarily upload one or more motifs. In Scansite 2.0, we now allow researchers to submit motifs directly into the Scansite database to make them available to other users. This should contribute favorably to the number and diversity of motif types that can be searched for in protein sequence queries. However, we cannot vouch for the accuracy of user-submitted motifs. To control for this, the web site allows users the option of including or rejecting user-submitted motifs in their scans. In addition, user-submitted motifs can be individually selected along with our standard Scansite motifs when using the Motif Scan programs. Interested users should contact us for information on adding motifs to the Scansite database.
Open source
Scansite 2.0 is a completely rewritten version of the original program, developed entirely at the Massachusetts Institute of Technology. We are releasing the source code for Scansite under the terms of the GNU General Public License, version 2 (Free Software Foundation, http://www.gnu.org/licenses/gpl.txt). Researchers interested in the fine details of our score calculations and other methodologies will thus have access to them and laboratories considering writing similar web applications can use our code to get started. The PSSMs for the 62 Scansite motifs, however, remain proprietary and are not included in the release. This policy is intended to prevent incorporation of the motifs into unauthorized commercial products. Use of the motifs on our public web site is permitted for all users, whether commercial or not. Anyone developing new features for Scansite is encouraged to submit changes back to us for inclusion in future public releases.
FUTURE DIRECTIONS
The revision of Scansite has produced a significantly faster and more efficient program for finding probable protein interactions. Focused searches enabled by incorporation of a relational database will help investigators target Scansite 2.0 to their own model organisms and experiments. New motifs will continue to be added to Scansite as they become available from oriented peptide library experiments. Researchers are encouraged to submit motifs of their own to our database for others to use. More specialized protein databases will be added over time, such as the RefSeq database and the mouse proteome. We are in the process of installing a second Scansite server for batch processing of long lists of sequences such as those obtained from DNA microarray experiments or genomic sequencing efforts. Future additions will include the ability to search among specific tissue types, the ability to adjust scores for predicted interactions based on their evolutionary conservation in orthologues and paralogues, the ability to restrict predicted interactions to proteins that co-localize in the same subcellular compartment, the ability to correlate predicted interactions with published data in the literature in an automated manner and the ability to automatically generate signaling network-style diagrams based on predicted interactions.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to acknowledge the work done by developers who contributed to Scansite version 1.0, especially German Leparc and Stefano Volinia, as well as to members of the Yaffe and Cantley laboratories that provided the experimental data and beta-tested the programs. This work was funded by a Merck Genome Research Institute grant, the Merck/MIT Collaboration Program, NIH grants GM-60594 (M.B.Y.), GM-56203 (L.C.C.) and GM-52981 (M.B.Y.) and a Burroughs-Wellcome Career Development Award to M.B.Y.
REFERENCES
- 1.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J. and Wheeler,D.L. (2003) GenBank. Nucleic Acids Res., 31, 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stoesser G., Baker,W., van den Broek,A., Garcia-Pastor,M., Kanz,C., Kulikova,T., Leinonen,R., Lin,Q., Lombard,V., Lopez,R. et al. (2003) The EMBL Nucleotide Sequence Database: major new developments. Nucleic Acids Res., 31, 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyazaki S., Sugawara,H., Gojobori,T. and Tateno,Y. (2003) DNA Data Bank of Japan (DDBJ) in XML. Nucleic Acids Res., 31, 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clamp M., Andrews,D., Barker,D., Bevan,P., Cameron,G., Chen,Y., Clark,L., Cox,T., Cuff,J., Curwen,V. et al. (2003) Ensembl 2002: accommodating comparative genomics. Nucleic Acids Res., 31, 38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bader G.D., Betel,D. and Hogue,C.W.V. (2003) BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res., 31, 248–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xenarios I., Fernandez,E., Salwinski,L., Duan,X.J., Thompson,M.J., Marcotte,E.M. and Eisenberg,D. (2001) DIP: The Database of Interacting Proteins: 2001 update. Nucleic Acids Res., 29, 239–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zanzoni A., Montecchi-Palazzi,L., Quondam,M., Ausiello,G., Helmer-Citterich,M. and Cesareni,G. (2002) MINT: a Molecular INTeraction database. FEBS Lett., 513, 135–140. [DOI] [PubMed] [Google Scholar]
- 9.Songyang Z., Shoelson,S.E., Chaudhuri,M., Gish,G., Pawson,T., Haser,W.G., King,F., Roberts,T., Ratnofsky,S. and Lechleider,R.J. (1993) SH2 domains recognize specific phosphopeptide sequences. Cell, 72, 767–778. [DOI] [PubMed] [Google Scholar]
- 10.Songyang Z., Blechner,S., Hoagland,N., Hoekstra,M.F., Piwnica-Worms,H. and Cantley,L.C. (1994) Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr. Biol., 4, 973–982. [DOI] [PubMed] [Google Scholar]
- 11.Yaffe M.B., Rittinger,K., Volinia,S., Caron,P.R., Aitken,A., Leffers,H., Gamblin,S.J., Smerdon,S.J. and Cantley,L.C. (1997) The structural basis for 14-3-3:phosphopeptide binding specificity. Cell, 91, 961–971. [DOI] [PubMed] [Google Scholar]
- 12.Songyang Z. and Cantley,L.C. (1998) The use of peptide library for the determination of kinase peptide substrates. Methods Mol. Biol., 87, 87–98. [DOI] [PubMed] [Google Scholar]
- 13.Yaffe M.B. and Cantley,L.C. (2000) Mapping specificity determinants for protein-protein association using protein fusions and random peptide libraries. Methods Enzymol., 328, 157–170. [DOI] [PubMed] [Google Scholar]
- 14.Kay B.K., Winter,J. and McCafferty,J. (1996) Phage Display of Peptides and Proteins: a Laboratory Manual. Academic Press, San Diego, CA. [Google Scholar]
- 15.Yaffe M.B., Leparc,G.G., Lai,J., Obata,T., Volinia,S. and Cantley,L.C. (2001) A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat. Biotechnol., 19, 348–353. [DOI] [PubMed] [Google Scholar]
- 16.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L.L. (2002) The Pfam Protein Families Database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letunic I., Goodstadt,L., Dickens,N.J., Doerks,T., Schultz,J., Mott,R., Ciccarelli,F., Copley,R.R., Ponting,C.P. and Bork,P. (2002) Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res., 30, 242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Obenauer J.C. and Yaffe,M.B. (2003) Computational prediction of protein–protein interactions. In Fu,H. (ed.) Protein–Protein Interactions: Methods and Protocols. Humana Press, Towata, NJ, in press. [Google Scholar]
- 19.Emini E.A., Hughes,J.V., Perlow,D.S. and Boger,J. (1985) Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J. Virol., 55, 836–839. [DOI] [PMC free article] [PubMed] [Google Scholar]