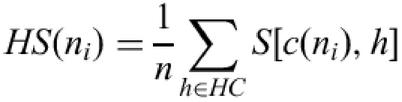Abstract
EUGÈNE'HOM is a gene prediction software for eukaryotic organisms based on comparative analysis. EUGÈNE'HOM is able to take into account multiple homologous sequences from more or less closely related organisms. It integrates the results of TBLASTX analysis, splice site and start codon prediction and a robust coding/non-coding probabilistic model which allows EUGÈNE'HOM to handle sequences from a variety of organisms. The current target of EUGÈNE'HOM is plant sequences. The EUGÈNE'HOM web site is available at http://genopole.toulouse.inra.fr/bioinfo/eugene/EuGeneHom/cgi-bin/EuGeneHom.pl.
INTRODUCTION
With the increasing number of sequenced organisms, exploiting sequence similarity information between homologous sequences has become a major challenge in the process of annotation and gene prediction (1). Sequence conservation information is largely organism independent and several gene prediction programs have been developed which are based on both exon conservation and splice site prediction information (2–5). The spectrum of application of these programs is wide and mainly limited by the splice-site model.
In order to improve gene prediction quality, more information should be taken into account. EUGÈNE'HOM has been developed to predict gene structures by combining conservation information with splice site prediction but also coding/non-coding statistics (6). Usual DNA level probabilistic models for coding/non-coding sequences are however extremely sensitive to the organism and GC% of the sequence and the introduction of such information strongly limits the scope of application of the software. To avoid this, EUGÈNE'HOM relies on an amino acid level probabilistic model which is largely organism independent and nevertheless improves gene prediction quality.
Useful information may also come from the variety of homologous sequences that may be taken into account to identify conserved exons. Because of evolution, each sequence may provide conservation (or divergence) information on different regions of the sequence. EUGÈNE'HOM has therefore been designed to take into account several homologous sequences simultaneously. A large spectrum of uses is possible, without any restriction to a particular evolutionary distance between the sequences considered.
Finally, to account for possible exon–intron structure evolution between distant sequences, EUGÈNE'HOM makes no assumption on the similarity of the gene structure (e.g. conservation of exon boundaries, exon order and repetitions) between the target sequence and the homologous sequences considered. Predictions can thus be performed using non-assembled shotgun sequences.
To perform the prediction itself, EUGÈNE'HOM uses the graph-based model and linear time dynamic programming algorithm of EUGÈNE (7), a gene prediction software for Arabidopsis thaliana designed to integrate arbitrary sources of evidence. This graph-model allows the prediction of several partial or complete genes on both strands and EUGÈNE'HOM inherits this ability.
The web site itself allows the user to perform gene prediction directly on a genomic sequence from one or more homologous DNA sequences (either genomic sequences or cDNA) and to compare it to an existing annotation provided in GFF format. Additionally, the user may enter specific information on signals and regions to either explore alternative predictions or to take into account further information that may help the prediction (e.g. repeats, EST matches, non canonical splice sites).
MATERIALS AND METHODS
Software description
As in all existing gene finders, a prediction in EUGÈNE'HOM is defined as a sequence of regions (exons coding in a specific frame, introns, UTR and intergenic regions) separated by signals and which follows the usual laws of gene structures: a gene starts with a 5′UTR, followed by a sequence of exons and introns, ending with a 3′UTR. Introns must be bordered by splice sites and the first and last exons respectively bordered by START and STOP codons. Exon assembly must respect the codon structure and should not contain in-frame STOPs. To choose a prediction among all possible ones, we give them a score defined as a sum of elementary scores: scores of the signals (splice sites, STOP and START codons) and scores of the nucleotides in the context of the predicted regions (coding in a specific frame, intronic on a specific strand or intergenic).
From all these scores, an optimal score prediction is computed in linear time using the dynamic programming algorithm of EUGÈNE (7).
Signal scores
Potential STARTs, STOPs and canonical AG/GT splice sites are detected and assigned a score. For STARTs and splice sites, the score is derived from a log-likelihood ratio computed using a first order weight array model (WAM) (8). Following the observation that splice sites are relatively well conserved within a taxonomic group (4), these models were built from expertised datasets of A.thaliana (9,10). If λ is the log-likelihood ratio obtained, the score S used is S=aλ+b where a and b are parameters.
Coding score
To characterise the coding/non-coding potential of a given region, we have used a two step probabilistic model. We consider that a coding sequence is first generated as a sequence of amino acids using a second order Markov model and then reverse translated using a uniform codon usage table. The Markov model was estimated from the SWISS-PROT database (11). For non-coding sequences, a zero-order Markov model is estimated on the sequence itself. The score of a nucleotide in a given region is defined as the logarithm of the probability of its emission in the corresponding probabilistic model.
Compared to classical DNA Markov models, this amino acid level Markov model provides less information but is also less sensitive to codon usage and GC%.
Homology scoring
The scoring system of EUGÈNE'HOM is based on the assumption that coding regions are more highly conserved than non-coding ones at the amino acid level. To detect conservations between sequences, EUGÈNE'HOM relies on an NCBI TBLASTX search (12) of the query sequence against the homologous sequences provided. To maximise both sensitivity and specificity, the following options are used: -F T (SEG filter activated), -e 1e-6 (threshold for expect value), -b 50000 (number of alignments). The -m 7 (XML format output) is also used to facilitate output parsing. The substitution matrices used are based on the usual PAM and BLOSUM matrices except that all the entries that correspond to STOP are modified to −500 in order to reduce false-positive hits.
The choice of an amino acid level comparison rather than a DNA level comparison (using e.g. BLASTN) was done to better accommodate homologous sequences from distant organisms. It should also improve discrimination between the different types of mutations in coding regions (that should favor amino acid conservation) and other conserved regions (which has no a priori reason to do so).
A local alignment algorithm was chosen in order to take into account the fact that gene structures may vary between species or even inside a given gene family: within a group of homologous genes, some protein domains may be shared by different pairs. Unlike global alignment methods that require extended pairwise conservation, a local alignment method allows identification of local similarities from multiple sequences.
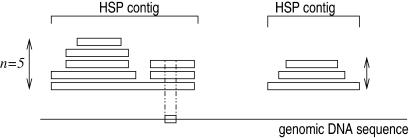
The TBLASTX search returns a set of high scoring pairs (HSP), each in a given frame. All pairs of HSP which overlap and are in the same frame are clustered together in so-called ‘HSP contigs’ (Fig. 1). To associate a homology score with a given nucleotide ni in the context of a coding region, we consider (if it exists), the single HSP contig HC that overlap the nucleotide in the sequence and which is in the same frame. Let n be the maximum number of HSPs in the cluster that overlap a single position. Let c(ni) be the codon that contains ni in the sequence, for each HSP h in the cluster that overlaps the codon c(ni), we define S[c(ni), h] to be the matrix substitution score for the amino acid coded by c(ni) in the HSP alignment. This score is considered as equal to zero for non-overlapping HSP. The homology score for the nucleotide in the context of the coding region considered is defined as:
Figure 1.
Two HSP contigs on the same frame above the genomic DNA sequence. The score used for each nucleotide in the codon visualized on the genomic sequence is equal to the sum of the elementary substitution scores of each HSP in the contig above divided by n, the maximum height of the contig.
This score is rescaled and added to the coding score.
Datasets
Beyond the previously cited datasets, we also used a set of 63 homologous plant sequences, mainly from A.thaliana (S. Aubourg, personal communication). The genes in this set come from five plant gene families (alcohol dehydrogenase, terpene synthase, cinnamyl alcohol dehydrogenase and two unknown function families). From each family of this set, we extracted the two pairs of sequences which respectively presented the highest and lowest identity at the amino acid level. These pairs of sequences were selected for evaluation purposes and the remaining sequences for parameter estimation. After careful examination of the annotation, we discarded 12 sequences because of inconsistent annotation or non-canonical splice sites. We finally obtained a test set of 16 genes organised in nine pairs (the same gene may appear in more than one pair) and a training dataset of 35 genes.
EUGÈNE'HOM behavior depends on a set of five parameters used to rescale each information source: two parameters for splice sites, two for START and STOP signals and one for the homology score. To estimate these parameters, we directly optimised the accuracy of EUGÈNE'HOM on the training data-set using a dedicated optimisation algorithm combining genetic algorithm and line search (7).
WEB SITE DESCRIPTION
Inputs
The first input required by EUGÈNE'HOM is the genomic sequence to be annotated. It can be provided either in FASTA format or as raw DNA (without any header). The sequence can be uploaded or pasted.
Then a set of supposedly homologous DNA sequences in multi-fasta format must be provided. This set can include genomic or cDNA sequences from different organisms. Ideally, it should not contain a genomic sequence which overlaps the query. The amino acid substitution matrix used to compute substitution scores should be selected here (default to BLOSUM80).
For expert users, an advanced query form is also available. In this form, the user may control the prediction by deactivating the use of the amino acid level coding model (keeping only homology scoring) or enforce the prediction of complete genes only. Also, the prediction can take into account specific user information using a simple language on signals and regions. For example, to explore alternative predictions, the user can delete a predicted donor site on the forward strand at position 877 by the simple sentence donor f 877 0.0 (see the web site or the Supplementary Material for more details).
The graphical and textual representation of the prediction can also be controlled by several parameters: it is possible, for example, to change the image resolution, or to restrict the graphical representation to a region of the sequence (for zooming). Optionally, an existing annotation for the analysed sequence can be provided in GFF format. This annotation will be displayed in the graphical output to facilitate comparison with the prediction.
Outputs
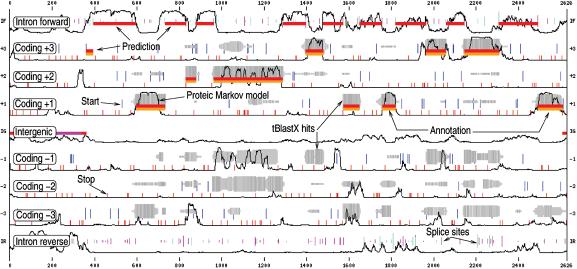
From these inputs, EUGÈNE'HOM produces a prediction (standard or GFF format) and a graphical output which represents this prediction, all the information used to compute it and the optional annotation. An example of EUGÈNE'HOM graphical output using the examples provided on the web site (‘Example’ buttons) is shown in Figure 2.
Figure 2.
An example of the graphical output of EUGÈNE'HOM: the sequence is on the x axis. The y axis corresponds to possible predictions. From top to bottom: intronic forward, frame 3, 2, 1 coding regions, then intergenic regions (IG) then frame -1, -2, -3 coding regions, then intronic reverse. In-frame START codons are represented as blue vertical lines. The longer the line, the better the score of the START. In-frame STOP codons are represented as small red vertical lines. Splice sites are visible on the intronic tracks as green and magenta vertical lines whose length indicates the site score. Thin black lines represent the smoothed normalised proteic coding/non coding score. HSP clusters are represented as grey blocks whose thickness is proportional to the number of hits at a given position and whose darkness is proportional to the homology score at this position. The GFF annotation provided by the user is visible as orange blocks. The prediction itself is visible as red blocks. In this example, available on the web site, EUGÈNE'HOM predicts a gene structure that matches the annotation.
RESULTS
We performed a preliminary comparison of EUGÈNE'HOM with SGP-1 (4), one of the most closely related similarity-based programs available on the web, GenScan (13), a purely ab initio gene finder and two other versions of EUGÈNE, named EUGÈNEAT and EUGÈNEAT+HOM.
GenScan was applied to the 16 test sequences using http://genes.mit.edu/GENSCAN.html with default options except for the organism which was set to A.thaliana.
SGP-1 takes two sequences as input and annotates both. It was therefore applied to the nine sequence test pairs using http://soft.ice.mpg.de/sgp-1/sgp-1noemail.html. For genes which appear in two pairs, we retained the best prediction obtained. Options were chosen to be consistent with EUGÈNE'HOM assumptions: taxonomic group=angiosperm, alignment method=TBLASTX, no post-processing, substitution matrix =Blosum80. For three sequences, SGP-1 required deactivation of the ‘no post processing’ flag to run and the ‘monotonic increasing unique’ option was selected.
EUGÈNEAT is the current version of EUGÈNE optimized for A.thaliana (7). It uses a species specific nucleic coding model instead of a generic protein model and external start and splice site detection software dedicated to A.thaliana. However, it does not exploit conservation information.
EUGÈNEAT+HOM is a combined version obtained by directly adding to EUGÈNEAT scores the proteic coding model and the homology scoring method of EUGÈNE'HOM.
Gene prediction accuracy was measured in terms of sensitivity and specificity both at the exon and nucleotide level (14). The results are presented in Table 1.
Table 1. Performances of the plant homology test set.
| Program | SNe | SPe | SNn | SPn | AC |
|---|---|---|---|---|---|
| SGP-1 | 11.48 | 35.00 | 35.29 | 93.02 | 0.53 |
| Genscan | 52.46 | 49.61 | 62.86 | 79.80 | 0.61 |
| EUGÈNEAT | 63.11 | 63.11 | 82.91 | 74.57 | 0.69 |
| EUGÈNE'HOM | 76.23 | 54.07 | 96.37 | 71.24 | 0.75 |
| EUGÈNEAT+HOM | 88.52 | 74.48 | 97.80 | 76.34 | 0.80 |
SNe, exon-level sensitivity; SPe, exon-level specificity; SNn, nucleotide-level sensitivity; SPn, nucleotide-level specificity; AC, approximate correlation (14). Gene-level accuracy was not computed because of the small size of the dataset, including a total of 16 genes, 122 exons and 75 kb.
The performances of SGP-1, GenScan and EUGÈNEAT are lower than previously reported (4,9). This is certainly due to the high number of exons composing the genes of this dataset (average of eight per gene). Moreover, some sequences come from other plant species than A.thaliana, thus disrupting the coding models of GenScan and EUGÈNEAT.
The comparison of the different versions of EUGÈNE confirms the accuracy improvement resulting from the addition of the new EUGÈNE'HOM methodologies.
Despite the limited size of the test set, these results show that without organism-specific coding models, EUGÈNE'HOM is able to predict the exon–intron structure of plant genes with a good accuracy. The construction of splice site models for vertebrates and mammals will further extend the scope of application of EUGÈNE'HOM.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Sébastien Aubourg from the Unité de Recherche en Génomique Végétale, INRA/CNRS for the set of homologous sequences, Laurent Bize from the Unité de Biométrie in Intelligence Artificielle, INRA for the XML Blast parser used in EUGÈNE'HOM web site and Richard Cooke from the University of Perpignan for polishing the english. This work is supported in part by GENOPLANTE, under project Bi2001049.
REFERENCES
- 1.Mathé C., Sagot,M.-F., Schiex,T. and Rouzé,P. (2002) Current methods of gene prediction, their strengths and weaknesses. Nucleic Acids Res., 30, 4103–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blayo P., Rouzé,P. and Sagot,M.-F. (2003) Orphan gene finding—an exon assembly approach. Theor. Comp. Sci., 290, 1407–1431. [Google Scholar]
- 3.Novichkov P., Gelfand,M. and Mironov,A. (2001) Gene recognition in eukaryotic DNA by comparison of genomic sequences. Bioinformatics, 17, 1011–1018. [DOI] [PubMed] [Google Scholar]
- 4.Wiehe T., Gebauer-Jung,S., Mitchell-Olds,T. and Guigo,R. (2001) SGP-1: prediction and validation of homologous genes based on sequence alignments. Genome Res., 11, 1574–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bafna V. and Huson,D. (2000) The conserved exon method for gene finding. Proc. Int. Conf. Intell. Syst. Mol. Biol., 8, 3–12. [PubMed] [Google Scholar]
- 6.Korf I., Flicek,P., Duan,D. and Brent,M. (2001) Integrating genomic homology into gene structure prediction. Bioinformatics, 17, S140–S148. [DOI] [PubMed] [Google Scholar]
- 7.Schiex T., Moisan,A. and Rouzé,P. (2001) Eugène, an eukaryotic gene finder that combines several type of evidence. In Computational Biology, selected papers from JOBIM'2000 number 2066 in LNCS, Springer Verlag, pp. 118–133. [Google Scholar]
- 8.Zhang M. and Marr,T. (1993) A weight array method for splicing signal analysis. Comput. Appl. Biosci., 9, 499–509. [DOI] [PubMed] [Google Scholar]
- 9.Pavy N., Rombauts,S., Déhais,P. Mathé,C., Ramana,D., Leroy,P. and Rouzé,P. (1999) Evaluation of gene prediction software using a genomic data set: application to Arabidopsis thaliana sequences. Bioinformatics, 15, 887–899. [DOI] [PubMed] [Google Scholar]
- 10.Korning P., Hebsgaard,S., Rouzé,P. and Brunak,S. (1996) Cleaning the GenBank Arabidopsis thaliana data set. Nucleic Acids Res., 24, 316–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M.C., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O'Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein knowledge base and its supplement TrEMBL. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul S., Madden,T., Schaffer,A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burge C. and Karlin,S. (1997) Prediction of complete gene structures in human genomic DNA. J. Mol. Biol., 268, 78–94. [DOI] [PubMed] [Google Scholar]
- 14.Burset M. and Guigo,R. (1996) Evaluation of gene structure prediction programs. Genomics, 34, 353–367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.