Abstract
We describe the current status of the Java molecular graphics tool, MolSurfer. MolSurfer has been designed to assist the analysis of the structures and physico-chemical properties of macromolecular interfaces. MolSurfer provides a coupled display of two-dimensional (2D) maps of the interfaces generated with the ADS software and a three-dimensional (3D) view of the macromolecular structure in the Java PDB viewer, WebMol. The interfaces are analytically defined and properties such as electrostatic potential or hydrophobicity are projected on to them. MolSurfer has been applied previously to analyze a set of 39 protein–protein complexes, with structures available from the Protein Data Bank (PDB). A new application, described here, is the visualization of 75 interfaces in structures of protein–DNA and protein–RNA complexes. Another new feature is that the MolSurfer web server is now able to compute and map Poisson–Boltzmann electrostatic potentials of macromolecules onto interfaces. The MolSurfer web server is available at http://projects.villa-bosch.de/mcm/software/molsurfer.
INTRODUCTION
Macromolecular interfaces, such as those between two proteins and or between protein and DNA or RNA molecules, can be very complex in shape, dynamics and physico-chemical properties. For studying the surfaces related to three-dimensional (3D) objects such as biomacromolecules, two-dimensional (2D) projections and maps are useful, because they can show the entire surface and the distribution of many properties simultaneously. Many parts of the surface that cannot be seen from a chosen viewpoint in 3D may be seen without difficulty in 2D projections. On the other hand, any projection of a 3D surface onto a flat 2D map introduces distortions and the 3D representation of a molecule and its surface is more accurate and realistic.
MolSurfer (1) was developed for visualization of 2D projections of interfaces as maps in such a way that they could be used to navigate through the 3D view of a macromolecular complex. It has been applied to studying the intermolecular interaction properties of proteins (1) and to the interactive analysis of the trajectories from molecular dynamics simulations of ligand–protein binding (2). We have now extended its capabilities to nucleic acids. MolSurfer facilitates the location of interfacial voids and ‘hot-spot’ patches of surface properties of interest and the identification of patterns in the distribution of interaction properties over the interfaces between macromolecules.
DESCRIPTION
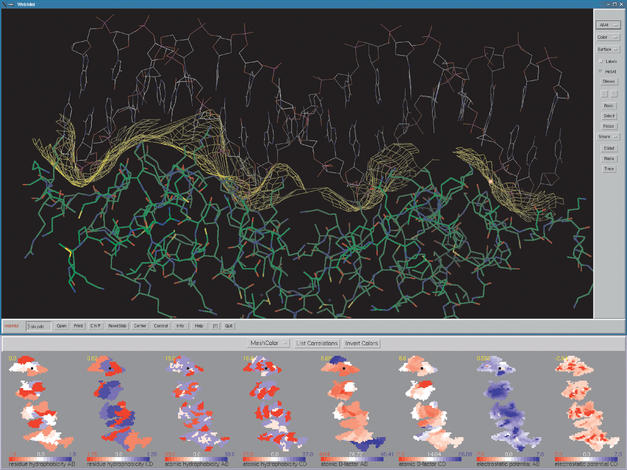
MolSurfer is a Java graphical tool that links a 2D projection of a macromolecular interface to a 3D view of the structure of the macromolecular complex. MolSurfer uses the representation of the macromolecular interface generated by ADSI, the Interface-mapping module of the ADS (Analytically Defined Surfaces) software (3), and WebMol, a Java-based PDB viewer (4). Technically, it consists of two linked windows (Fig. 1).
Figure 1.
A screen snapshot from a session in which MolSurfer is used to analyze the features of the interaction between the glucocorticoid receptor DNA binding domain (chains A and B of the PDB entry 1glu) and a DNA fragment (chains C and D). The distributions of atomic and residue hydrophobicity and electrostatic potential on the two molecules are displayed on the 2D maps of the molecular interface. Electrostatic complementarity between the negatively charged DNA (red) and the positively charged protein (blue) is readily apparent while hydrophobic complementarity is only partial. The mouse position on any of the maps is simultaneously seen on the other 2D maps (black squares) and on the interface in the 3D view (a spot, coloured by map property, red in this case).
The first window, MolSurfer, displays 2D maps of molecular properties computed with ADSI. The following properties are mapped: (i) distance between the closest atoms of the two molecules; (ii) distance between the van der Waals surfaces of the two molecules; (iii) hydrophobicity of the closest residues of each molecule; (iv) hydrophobicity of the closest atoms of each molecule; (v) crystallographic temperature factor of the closest atoms of each molecule; (vi) electrostatic potential from each molecule (optional); (vii) user-defined property assigned to atoms (optional). Properties (i) and (ii) describe the degree of steric complementarity between the macromolecules forming a complex. Properties (iii) and (iv) may be used to estimate the distribution of desolvation free energy changes upon complexation of the macromolecules (5,6). Property (v) is a measure of the flexibility of the interacting molecules and property (vi), computed by the program UHBD (7), is an indicator of the strength of charge–charge interactions. One possible choice of the property (vii) is the change in the binding free energy when the residue closest to the interface point in a protein is mutated to alanine. Such information permits direct identification of the important energetic ‘hotspots’ in the interface (8).
The second window, WebMol, displays the molecules in 3D. The two windows are linked in such a way that mouse manipulations on the 2D map window change the map and the 3D view. This link makes it possible to use the 2D maps to navigate in the 3D view. The interface is shown via triangulation on the 3D view and can be coloured in different ways. Two-dimensional maps have variable colour scaling to facilitate analysis of the distribution of atomic properties. Cross-correlation coefficients of different maps can be computed allowing the detection of correlations across the interface.
MolSurfer is suitable for performing in-depth studies of individual pairs of molecules and for large-scale analyses of sets of structures, such as databases of diverse protein–protein and protein–DNA/RNA complexes or a set of mutants of one macromolecular complex. The required input data for using MolSurfer are files containing the atomic coordinates of each molecule in Protein Data Bank (PDB) (9) format. The MolSurfer representation of macromolecular interactions may readily be used in web publishing. For this, locations of interest should be described or labeled on the 2D map so that the user can easily find the location in the 3D view in order to carry out further investigations using the numerous analysis and graphical features implemented in WebMol. Stand-alone and applet versions of MolSurfer are available, allowing MolSurfer to be accessed using the Java run-time system or directly using Java Development Kit and Java-enabled web browsers, respectively. Complete documentation of MolSurfer may be found at the URL http://projects.villa-bosch.de/mcm/software/molsurfer. At this URL, users can download MolSurfer or access a web server for generating a MolSurfer representation of the interfaces of user-specified macromolecules.
A new feature of the current version of the MolSurfer web server is that it can compute the electrostatic potentials of the individual macromolecules and project them onto the interface. This allows a detailed study of the electrostatic complementarity on macromolecular interfaces (10). For this feature, the user should supply the macromolecular coordinates along with atomic charges and radii in PQR format. Atomic charges and radii can be added to a PDB coordinate file to generate a PQR format file using, for example, the PDB2PQR web portal, http://nbcr.sdsc.edu/pdb2pqr/ (11). The electrostatic potentials of the macromolecules are computed by numerically solving the finite-difference Poisson–Boltzmann equation using the program UHBD (7). These computations give values of the electrostatic potentials on 3D grids, which are then interpolated onto the points of the macromolecular interface. A grid focusing procedure is applied such that the entire macromolecule is required to be inside the first grid and only the interface points are required to be inside the final grid. The electrostatic potential at the boundary of the final grid is interpolated from the first grid. This focusing procedure ensures adequate resolution of the final grid for interpolation and a fast and accurate description of the electrostatic potentials for typical protein–protein and protein–DNA/RNA interfaces using grids having dimensions 653. Finally, projection of the grid electrostatic potentials to the interface points is done using the UHBD commands for interpolation of potentials. In the Poisson–Boltzmann description, the protein interior (defined either by the van der Waals surfaces of the atoms or as a molecular surface obtained by rolling a spherical solvent probe) has a lower dielectric permittivity than the solvent. The solvent can be modeled under different ionic strength conditions, the ionic solvent is usually located outside the Stern layer, defined by the radius of the ions (7). The related parameters can be changed by the user in the MolSurfer map request page.
APPLICATION
As well as application to explore protein–ligand trajectories from molecular dynamics simulations (2), the application of MolSurfer to the analysis of interactions in protein–protein and protein–DNA/RNA complexes is demonstrated at the MolSurfer URL. Two representative datasets, of 39 protein–protein complexes (1) and 75 protein–DNA/RNA complexes (12), are presented. These datasets demonstrate important features of macromolecular interfaces that are easily visualized with MolSurfer. For example, the discontinuous and holely nature of many interfaces is observed, with more pronounced discontinuities in protein–DNA/RNA interfaces compared to protein–protein interfaces. Electrostatic interactions and electrostatic complementarity are significantly stronger at protein–DNA/RNA interfaces than at protein–protein interfaces. Hotspots of particularly important residues that make strong macromolecular interactions are different for these two classes of interface. Protein–protein interfaces often show hydrophobic complementarity in hotspot regions of interaction. Protein–DNA/RNA interfaces do not show such complementarity; on the contrary, the residues and atoms of proteins contacting nucleic acids are, as a rule, hydrophilic. There is, however, a tendency for clusters of ‘hydrophobic’ nucleic acids [A, T and U (6)] to be located in the central parts of the interfaces.
As well as highlighting these characteristic differences between different interface types, MolSurfer can be used for in-depth analysis of particular macromolecular complexes. For example, areas of electrostatic mismatch or poor shape complementarity in a protein–protein/DNA/RNA interface might be located in the 2D maps and the adjacent residues identified and studied, by linking with the 3D view window, as candidates for site-directed mutagenesis. A tutorial example of how to explore the electrostatic and hydrophobic complementarity of a protein–protein interface is given at the MolSurfer webserver.
Acknowledgments
ACKNOWLEDGEMENT
Financial support from the Klaus Tschira Foundation is gratefully acknowledged.
REFERENCES
- 1.Gabdoulline R.R., Wade,R.C. and Walther,D. (1999) MolSurfer: two dimensional maps for navigating three-dimensional structures of proteins. Trends Biochem. Sci., 24, 285–287. [DOI] [PubMed] [Google Scholar]
- 2.Luedemann S.K., Gabdoulline,R.R., Lounnas,V. and Wade,R.C. (2001) Substrate access to cytochrome P450cam investigated by molecular dynamics simulations: an interactive look at the underlying mechanisms. Internet. J. Chem., 4, 6 (www.ijc.com/articles/2001v4/6/). [Google Scholar]
- 3.Gabdoulline R.R. and Wade,R.C. (1996) Analitically defined surfaces to analyze molecular interaction properties. J. Mol. Graph., 14, 341–353. [DOI] [PubMed] [Google Scholar]
- 4.Walther D. (1997) WebMol—a Java based PDB viewer. Trends Biochem. Sci., 22, 274–275. [DOI] [PubMed] [Google Scholar]
- 5.Eisenberg D., Wesson,M. and Yamashita,M. (1989) Interpretation of protein folding and binding with atomic solvation parameters. Chem. Scripta, 29A, 217–221. [Google Scholar]
- 6.Shih P., Pedersen,L.G., Gibbs,P.R. and Wolfensen,R. (1998) Hydrophobicities of the nucleic acid bases: distribution coefficients from water to cyclohexane. J. Mol. Biol., 280, 421–430. [DOI] [PubMed] [Google Scholar]
- 7.Madura J.D., Briggs,J.M., Wade,R.C., Davis,M.E., Luty,B.A., Ilin,A., Antosiewicz,J., Gilson,M.K., Bagheri,B., Scott,L.R. and McCammon,J.A. (1995) Electrostatic and diffusion of molecules in solution: simulations with the University of Houston Brownian Dynamics program. Comp. Phys. Comm., 91, 57–95. [Google Scholar]
- 8.Bogan A.A. and Thorn,K.S. (1998) Anatomy of hot spots in protein interfaces. J. Mol. Biol., 280, 1–9. [DOI] [PubMed] [Google Scholar]
- 9.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCoy A.J., Chandana,E.V. and Colman,P.M. (1997) Electrostatic complementarity at protein/protein interfaces. J. Mol. Biol., 268, 570–584. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen J.E., Andersen,K.V., Honig,B., Hooft,R.W., Klebe,G., Vriend,G. and Wade,R.C. (1999) Improving macromolecular electrostatics calculations. Protein Eng., 12, 657–662. [DOI] [PubMed] [Google Scholar]
- 12.Nadassy K., Wodak,S.J. and Janin,J. (1999) Structural features of protein-nucleic acid recognition sites. Biochemistry, 38, 1999–2017. [DOI] [PubMed] [Google Scholar]