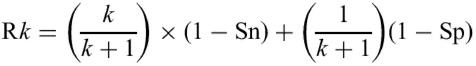Abstract
AMIGene (Annotation of MIcrobial Genes) is an application for automatically identifying the most likely coding sequences (CDSs) in a large contig or a complete bacterial genome sequence. The first step in AMIGene is dedicated to the construction of Markov models that fit the input genomic data (i.e. the gene model), followed by the combination of well-known gene-finding methods and an heuristic approach for the selection of the most likely CDSs. The web interface allows the user to select one or several gene models applied to the analysis of the input sequence by the AMIGene program and to visualize the list of predicted CDSs graphically and in a downloadable text format. The AMIGene web site is accessible at the following address: http://www.genoscope.cns.fr/agc/tools/amigene/index.html (Contact: sbocs@genoscope.cns.fr).
INTRODUCTION
Intrinsic methods for predicting coding regions extract information on gene locations using statistical patterns inside and outside gene regions as well as patterns typical of the gene boundaries. Several highly accurate prokaryotic gene-finding methods are based on Markov model algorithms [i.e. GeneMark (1) and Glimmer (2)]. The accuracy of these systems depends on models of protein coding regions (and non-coding regions in the case of GeneMark) derived either from experimentally validated training sets or from large amounts of anonymous DNA sequences. Identification of putative genes is followed by an examination of overlaps between selected ORFs (open reading frames) in order to eliminate doubtful candidates. In the context of bacterial genome annotation, we have intensively used these methods and compared their results using the graphical interface provided by Imagene (3). This interface allows one to superimpose results obtained by different strategies and/or gene-finding models and is very useful for pinpointing interesting features such as the coding sequences (CDSs) located at positions in which the coding prediction is good. We have noticed that although most of the predicted genes are identical, the Glimmer method tends to select additional ‘suspect’ CDSs (false positives). We also found examples in which Glimmer proposed a sequence on the opposite strand of the GeneMark prediction (4). Conversely, many short genes seem not to be identified by the GeneMark method and genes which are ‘atypical’ in their pattern of codon usage (compared to the average codon bias of the genome) could be missed (false negatives).
Using appropriate gene models with a coding prediction program such as GeneMark (1), the CDS selection is manually performed by keeping the longest CDSs which have a good coding prediction, a minimum overlap with adjacent CDSs (except in the case of frameshift detection) and maximum coverage of the nucleic sequence. These observations led us to mimic the behaviour of the expert in the AMIGene method to automatically identify the most likely CDSs in a large contig or a complete bacterial genome. Although AMIGene remains relatively similar to most existing gene finding systems, it is able to give more accurate predictions in some cases (4–6). The web interface described in this paper allows the user to run AMIGene on a raw DNA sequence, either with suitable gene models we have previously defined on several bacterial genomes, or with a new gene model computed from the user's input genomic data (http://www.genoscope.cns.fr/agc/tools/amigene/index.html).
METHODS
Generating the models for gene-finding
Running the AMIGene method requires the construction of Markov models that fit well with the input genomic data. A preliminary and essential step for a new genome annotation (anonymous DNA sequence) or a re-annotation process of an available prokaryotic genome (5) consists in the construction of appropriate gene models. To achieve this goal, two programs similar to the ones of M. Borodovsky [MakeMat (unpublished) and GeneMark (1)] have been developed (A. Viari, personal communication): (i) prokov-learn which uses inhomogeneous three-periodic Markov chain models of protein-coding regions along with ordinary Markov models of non-coding DNA sequences to build gene models; (ii) prokov-curve that incorporates these models into a Bayesian algorithm and analyzes DNA sequences locally within a sliding window (1).
The first procedure thus requires as input a set of predicted CDSs or a set of previously annotated genes. In the case of available complete bacterial genomes, the set of annotated genes has been extracted from the International Nucleotide Sequence Database (INSD: DDBJ/EMBL-EBI/GenBank). In the case of an anonymous bacterial sequence, we first search for the longest ORFs and the prokov-learn program is used to determine parameters of the Markov model of the protein-coding region (pre-matrix). Depending on the length of the input DNA sequence, the order of the Markov model is equal to two, three or four. For a Markov model of a non-coding sequence, we used the zero order model with four probability parameters estimated by genome specific frequencies of mononucleotides. These pre-matrices are then used in the AMIGene program for predicting coding regions in the original genomic sequence (set of predicted CDSs).
However, when the models are constructed by training on the bulk set of protein-coding genes, programs such as GeneMark (1) and Prokov-curve become insensitive to genes of minor inhomogeneity classes. For the Escherichia coli genome, whose genes have been divided into three classes that differ in codon usage pattern (7), class-specific models of protein-coding regions have improved the performance of the GeneMark method (8). Thus, our group systematically investigates codon usage differences using the multivariate statistical technique of factorial correspondence analysis (FCA) to identify major trends within the data set, i.e. annotated genes or predicted CDSs (9). The k-means clustering algorithm is also employed on relative synonymous codon usage (RSCU) values of the coding sequences (10), k being equal to 2, 3 or 4, depending on the major trend of the codon usage bias. For example, k=2 for bias due to genes lying on the leading versus lagging strands in the bacterial chromosome (11,12) and k=3 in the case of bacterial genomes for which recent horizontal gene transfer has occurred (7,13). The gene classes define the training sets for protein-coding regions and the rest of the sequence is included in the non-coding training set. Corresponding gene models are generated by the prokov-learn program (two, three or four in total) and subsequently used in the core of the AMIGene method. Several illustrations of this step are given on the AMIGene web site (http://www.genoscope.cns.fr/agc/tools/amigene/html/Method.html#1).
AMIGene: an heuristic to select the most likely CDSs
Given the sequence of a complete genome, we first look for the maximal CDSs, i.e. maximal segments in-frame between start and stop codons in the six reading frames. The putative CDSs >60 bp are retained. Then, the Prokov-curve method (A.Viari and J.Romanet, personal communication) uses the specific models built for the gene classes of the genome in parallel (see above), in order to compute the average coding probability of each identified CDS. AMIGene indicates the model (matrix 1, 2 or 3) that fits best in terms of coding probability. In order to accelerate the following steps, CDSs are first selected according to their coding probability. Two probability thresholds, prob-PC and sure-PC, are defined (Table 1). If the value of the coding probability is: (i) below the prob-PC threshold, the corresponding CDS is eliminated; (ii) between the prob-PC and sure-PC thresholds, the corresponding CDS is stored in a list containing the probable CDSs if its length is longer than the Prob-LMin threshold (Table 1); and (iii) above the sure-PC threshold, the corresponding CDS is stored in a list containing the sure CDSs. Therefore we defined an heuristic to select CDSs in order to take into account ambiguous choices between two overlapping CDSs and/or the presence of frameshifts in the DNA sequence. In order to avoid overlaps generated by the choice of the left-most start codon, AMIGene searches, when necessary, for an alternative start codon fitting well with the beginning of the coding prediction curve. Then the selection of the most likely CDSs consists of the elimination of false positives according to overlapping criteria between adjacent CDSs (AMIGene parameters are listed in Table 1); these overlaps may be either total (inclusion) or partial. The AMIGene method is divided into three main steps which are precisely described on our web site (http://www.genoscope.cns.fr/agc/tools/amigene/html/Method.html#2).
Table 1. Definition and value of the AMIGene parameters for three reference genomes (optimization process).
| Abbreviation | Definition | BACSU | ECOLI | MYCTU |
|---|---|---|---|---|
| Sure-Pc | Coding probability above which a CDS is interpreted as a sure CDS | 0.67 | 0.62 | 0.47 |
| Prob-Pc | Between the Sure-Pc and Prob-Pc thresholds a CDS is interpreted as a probable CDS | 0.35 | 0.40 | 0.21 |
| Prob-LMin | Minimum length (bp) of a probable CDS selected | 114 | 141 | 219 |
| Sure-ss-I | Minimum inclusion percentage between two sure CDSs transcribed on the same strand | 5 | 5 | 20 |
| Sure-os-I | Minimum inclusion percentage between two sure CDSs transcribed on the opposite strands | 30 | 56 | 70 |
| Sure-prob-O | Maximum overlapping percentage between a sure and a probable CDSs transcribed on the opposite strands | 5 | 5 | 37 |
| Prob-glob-IO | Maximum global score (%), including both inclusion and overlapping situations, between a probable CDS and all the other probable CDSs which partially or completely overlap this CDS | 86 | 75 | 99 |
ECOLI=Escherichia coli K12; BACSU=Bacillus subtilis 168; MYCTU=Mycobacterium tuberculosis H37Rv.
Setting the threshold of the parameter values
The threshold values used in the AMIGene method were first determined empirically, based on the examination of results obtained with several AMIGene runs on various bacterial genomes. Then a statistical validation of the chosen threshold values was performed using curated annotations from three bacterial genomes: E.coli K12 (50.8% GC-content), Bacillus subtilis 168 (43.5% GC-content) and Mycobacterium tuberculosis H37Rv (65.6% GC-content). E.coli annotated genes were extracted from the last update of the EcoGene data base (14) and we have used curated data from the SubtiList database (15) and from the TubercuList database (16). For each genome, we first defined three gene classes according to their codon usage (see below), which were subsequently used to build three gene models. Predictions made by the AMIGene program were compared to the corresponding annotation reference set. An AMIGene prediction was assumed to be correct if the predicted stop codon of the CDS matched the annotated stop. The sensitivity, Sn, is thus defined as the ratio of the number of correctly predicted genes to the number of genes annotated in the corresponding curated database. The specificity, Sp, is the ratio of the number of correctly predicted genes to the total number of genes predicted by AMIGene. Optimization of the AMIGene parameter values was performed by searching for the set of values that minimize the following risk Rk function:
where k is the penalization factor between false-negative and false-positive predictions. Successive minimizations in each direction (seven parameter values, Table 1) were performed on several iterations until the Rk variation between two successive iterations was <0.1% [i.e. Ri−(Ri+1)/Ri<0.1%]. This optimization process was performed for k=10 (false-negative predictions are penalized 10 times more than false-positive predictions) and led to three sets of parameter values for the reference genomes (Table 1). The AMIGene average accuracy of gene finding is then characterized by 98.3% sensitivity and 92.4% specificity. Based on the learning set, such values are probably an upper limit of the accuracy of AMIGene.
The accuracy of our method was then tested on bacterial genomes closely related to our models: Bacillus halodurans, E.coli O157:H7 and M.tuberculosis CDC1551. Annotation data were extracted from the INSD and compared to the set of AMIGene predicted CDSs, using the three gene models built for these genomes and the optimized parameter values of each related organism (Table 1). AMIGene predictions were very good for the B.halodurans genome (Sn=98.6% and Sp=89%) and the E.coli O157:H7 genome (Sn=96.8% and Sp=93.2%). Concerning the M.tuberculosis CDC1551 genome, the sensitivity was 94.7% with 85% specificity. This result is somewhat surprising since the two M.tuberculosis strains (H37Rv and CDC1551) share >90% identity at the DNA level. Additional statistical tests have been used to demonstrate that this lower prediction emerged from heterogeneity between the different sets of M.tuberculosis annotations (H37Rv and CDC1551; not shown).
The AMIGene web site
AMIGene is implemented in the C language and is available upon request as a stand-alone application or via a web server at the following URL: http://www.genoscope.cns.fr/agc/tools/amigene/index.html.
The home page of our software allows users to choose the AMIGene input parameters and is divided into four main sections. A precise description of this page can be found at the following URL: http://www.genoscope.cns.fr/agc/tools/amigene/html/helpForm.html; the ‘Gene Model’ section allows the user to either select existing matrices that have been computed on several bacterial gene classes or to build a new gene model (see below). In the latter case, the minimum recommended length of the input sequence is 10 kb. The ‘AMIGene parameters’ section allows the user to either choose the parameter values that have been optimized for three reference genomes with different GC contents (i.e. B.subtilis for a low GC%, E.coli for a medium GC% and M.tuberculosis for a high GC%; see below), or to define his/her own parameter values. It is however recommended to carefully read the detailed description of the heuristic we have implemented in AMIGene (http://www.genoscope.cns.fr/agc/tools/amigene/html/Method.html#2). The proposed default values are close to those obtained after the optimization procedure on the E.coli genome (see below). The third section (‘Sequence’), allows the user to enter a DNA sequence (either a large contig or a complete bacterial genome) and to choose the adapted genetic code. In the last section (‘Options’), several additional functions are proposed such as the translation of the predicted genes (leading to a Fasta file format which can be downloaded from the results home page) or the search for putative frameshifts using our ProFED method (16).
The home page of the AMIGene results includes the list of predicted CDSs in text format and a graph representing the protein coding potentials (both CDS positions and coding prediction curves in the six reading frames). The map is fully dynamic and allows the user to navigate along the genome (or contig) while the corresponding list of predicted CDSs is updated accordingly. The predicted CDSs are drawn using only the left-most start position on the sequence. The positions of putative frameshifts are also clearly indicated on this map and the nucleic or peptidic sequence of each predicted CDS can be retrieved independently. Finally this home page includes several files which can also be downloaded: two files containing predicted nucleic and protein sequences and one file containing the positions of the putative frameshifts (ProFED results; 16). More details on the AMIGene results page are available at the following URL: http://www.genoscope.cns.fr/agc/tools/amigene/html/helpViewer.html.
CONCLUSIONS
The AMIGene method, together with the web software presented here, has already been used to analyze >30 complete prokaryotic genomes and its gene-finding accuracy was assessed by comparison with existing annotations (5). Several interesting discrepancies were in favour of a better selection of CDSs using the biological heuristic developed in AMIGene. Although an alternative start codon is sometimes used in the CDS selection step, our method is not yet suitable for identifying true translation initiation sites. If an alternative start codon is proposed in the AMIGene output file, this only indicates that probably the left-most start codon is not correct. The next version of AMIGene will include correct gene start predictions, taking into account overlaps between adjacent CDSs, coding prediction curves, translation signals such as the Ribosome Binding Site, together with results of similarities in the protein databanks.
The use of two, three or four gene models instead of only one is clearly an improvement in the final selection. Whatever the reasons for intragenomic variations (e.g. codon bias, base content), the construction of several gene classes based on codon usage leads to Markov models that can uncover small genes which are difficult to spot using the typical model. Genes with atypical composition are candidates for being horizontally transferred genes, although additional evidence would be necessary to confirm this hypothesis (17,18). Indeed, identifying the number of gene classes based on their codon usage (with FCA and clustering statistical methods; see below) is more pertinent when performed by human experts. This work is currently under development using the GenoStar platform (19) (GenoAnnot and GenoBool modules; http://www.genostar.org) and deals with pitfalls which arise from the use of correspondence analysis in codon usage studies; the transformation performed on the original data (i.e. the absolute codon frequencies) decreases the amount of information and may introduce new biases (20). In addition to gene models currently available for 12 genomes, we plan to regularly add new gene models in our AMIGene web site, computed based on the codon usage analysis of other bacterial genomes.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by the French Centre National de la Recherche Scientifique (CNRS-URA8030) and the GENOPOLE of Evry. We thank Antoine Danchin (Genetics of Bacterial Genomes, Pasteur Institute) and Hugues Roest Crollius (Genoscope/CNRS-UMR8030, Evry) for their critical comments and suggestions.
REFERENCES
- 1.Borodovsky M. and McIninch,J.D. (1993) GeneMark: parallel gene recognition for both DNA strands. Comp. Chem., 17, 123–133. [Google Scholar]
- 2.Salzberg S.L., Delcher,A.L., Kasif,S. and White,O. (1998) Microbial gene identification using interpolated Markov models. Nucleic Acids Res., 26, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Médigue C., Rechenmann,F., Danchin,A. and Viari,A. (1999) Imagene: an integrated computer environment for sequence annotation and analysis. Bioinformatics, 15, 2–15. [DOI] [PubMed] [Google Scholar]
- 4.Médigue C., Wong,B.C.Y., Lin,M.C.M., Gu,Q., Bocs,S. and Danchin,A. (2002) The secE gene of Helicobacter pylori. J. Bacteriol., 184, 2837–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bocs S., Danchin,A. and Médigue,C. (2002) Re-annotation of genomes microbial CoDing Sequences: finding new genes and inaccurately annotated genes. BMC Bioinformatics, 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camus J.C., Pryor,M.J., Médigue,C. and Cole,S. (2002) Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology, 148, 2967–2973. [DOI] [PubMed] [Google Scholar]
- 7.Médigue C., Rouxel,T., Vigier,P., Hénaut,A. and Danchin,A. (1991) Evidence for horizontal gene transfer in Escherichia coli speciation. J. Mol. Biol., 222, 851–856. [DOI] [PubMed] [Google Scholar]
- 8.Borodovsky M., McIninch,J., Koonin,E., Rudd,K., Médigue,C. and Danchin,A. (1995) Detection of new genes in the bacterial genome using Markov models for three gene classes. Nucleic Acids Res., 23, 3554–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill M.O. (1974) Correspondence analysis: a neglected multivariate method. Appl. Stat., 23, 340–354. [Google Scholar]
- 10.Cover T.M. and Thomas,J.A. (1991) Elements of Information Theory. John Wiley, New York. [Google Scholar]
- 11.McInerney J.O. (1998) Replicational and transcriptional selection on codon usage in Borrelia burgdorferi. Proc. Natl Acad. Sci. USA, 95, 10698–10703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rocha E.P., Danchin,A. and Viari,A. (1999) Replication biases in bacteria. Mol. Microbiol., 32, 11–16. [DOI] [PubMed] [Google Scholar]
- 13.Moszer I., Rocha,E.P.C. and Danchin,A. (1999) Codon usage and lateral gene transfer in Bacillus subtilis. Curr. Opin. Microbiol., 2, 524–528. [DOI] [PubMed] [Google Scholar]
- 14.Rudd K.E. (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res., 28, 60–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moszer I., Jones,L.M., Sandrine, Moreira,C. and Danchin,A. (2002) SubtiList: the reference database for the Bacillus subtilis genome. Nucleic Acids Res., 30, 62–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Médigue C., Rose,M., Viari,A. and Danchin,A. (1999) Detecting and analysing sequencing errors: toward a high quality of the Bacillus subtilis genome sequence. Genome Res., 9, 1116–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang B. (2001) Limitations of compositional approach to identifying horizontally transferred genes. J. Mol. Evol., 53, 244–250. [DOI] [PubMed] [Google Scholar]
- 18.Koski L.B., Morton,R.A. and Golding,B. (2001) Codon bias and base composition are poor indicators of horizontally transferred genes. Mol. Biol. Evol., 18, 404–412. [DOI] [PubMed] [Google Scholar]
- 19.Durand P., Médigue,C., Morgat,A., Vandenbrouck,Y., Viari,A. and Rechemmann,F. (2003) Integration of data and methods for genome analysis. Curr. Opin. Drug Disc. Devel., 6, 346–352. [PubMed] [Google Scholar]
- 20.Perrière G. and Thioulouse,J. (2002) Use and misuse of correspondence analysis in codon usage studies. Nucleic Acids Res., 30, 4548–4555. [DOI] [PMC free article] [PubMed] [Google Scholar]