Abstract
iSPOT (http://cbm.bio.uniroma2.it/ispot) is a web tool developed to infer the recognition specificity of protein module families; it is based on the SPOT procedure that utilizes information from position-specific contacts, derived from the available domain/ligand complexes of known structure, and experimental interaction data to build a database of residue–residue contact frequencies. iSPOT is available to infer the interaction specificity of PDZ, SH3 and WW domains. For each family of protein domains, iSPOT evaluates the probability of interaction between a query domain of the specified families and an input protein/peptide sequence and makes it possible to search for potential binding partners of a given domain within the SWISS-PROT database. The experimentally derived interaction data utilized to build the PDZ, SH3 and WW databases of residue–residue contact frequencies are also accessible. Here we describe the application to the WW family of protein modules.
INTRODUCTION
Protein–protein interactions are of particular interest as they play an important role in regulating several biological processes within the cell, such as metabolic pathways, progression through cell cycle, protein synthesis and DNA replication (1). Families of protein modules often mediate protein–protein interactions by recognizing short sequence motifs within their target proteins. The ability to infer the recognition specificity of such families would help to increase our knowledge of protein interaction networks. Many computational methods can be used to infer protein interactions when interaction data are available in the form of binding peptide lists: regular expressions, which can be used to scan a protein sequence or a database of protein sequences to identify potential partners of interaction, and the Position Specific Scoring Matrices (PSSMs) (2,3). To this aim, specific tools such as PatMatch at the SGDTM (Saccharomyces Genome Database) (4), ScanProsite (5) or profile search software (2,6) can be used. Different methodologies can be applied when the domain and/or the domain/ligand complex of known structures are available, such as homology modelling and protein docking based methods (7,8) or Virtual Interaction Profile (VIP) (9).
The SPOT procedure (10) has been developed to infer the recognition specificity of families of protein modules. Given a family of protein modules, the analysis of all the complexes of known structure available in the Protein Data Bank (PDB) between a member of the family and one ligand allows the identification of contact residues in the binding surface.
The defined residue–residue interaction networks between the domain and its ligand, together with the experimentally-derived interaction data, can be used to define a database of frequencies of residue–residue contact pairs in established contact positions in the binding surface of stable experimental complexes. The procedure evaluates the probability of interaction between a domain and a peptide sequence with a score, corresponding to the sum of the residue pair frequencies in the domain-specific database. Thus, the prediction is based on the assumption that the interaction between two proteins can be described, in first approximation, as the sum of independent interactions between their contacting residues. Therefore the experimental information on interaction, contained at the level of domain and peptide sequence, is transferred by SPOT at the level of interacting residues. As a matter of fact, this transfer of information from whole sequence interacting pairs to residue interacting pairs makes it possible to infer the specificity of interaction for domain sequences of unknown structure and specificity on condition that at least some contact residues are identical to the ones observed in stable complexes.
The ability to infer the interaction specificity of protein modules whose interaction data are not available is a feature that makes iSPOT (11) unique in the landscape of the available tools for protein interaction prediction.
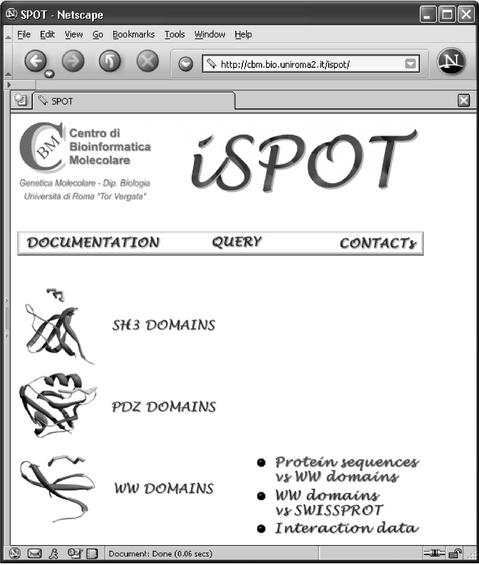
From the iSPOT home page (Fig. 1), the selection of the protein domain of interest can be operated among the SH3, PDZ and WW families. For each domain family, three links are available: (i) query protein sequence versus one or more domain sequences; (ii) single domains versus the SWISS-PROT database; and (iii) interaction data.
Figure 1.
iSPOT home page (http://cbm.bio.uniroma2.it/ispot). iSPOT features are accessible for each family of protein modules by clicking on the name of the family.
We describe here, as an example, the use of the iSPOT web tool for WW domains.
PROTEIN SEQUENCE VERSUS ONE OR MORE WW DOMAIN SEQUENCES
The protein query sequence has to be entered in the proper box of the WW input form in FASTA format or as plain text without blank characters (Fig. 2). A SWISS-PROT–TrEMBL accession code can also be provided. Input sequences are expected to contain only one-letter code residues as described in the documentation available in the web pages. The user can select one or more WW domain sequences from the list reporting the name of all WW domain sequences contained in the Pfam database version 7.8 (12). The protein sequence is scanned at one-residue steps with an eight-residue-long window. A ranked list of WW domains is then reported according to their evaluated propensity to bind any peptide belonging to the input sequence (Fig. 3). The score is normalized on the value of the best peptide for the given WW domain, this being obtained considering, for each peptide position, the best ranking residue in the WW-specific database of frequencies of interacting residues (10). Scores <0.5 are not listed in the output. The output list produced in the ‘one sequence’ mode (when a single protein sequence is submitted for prediction versus one or more domains) shows all the peptides of the query protein ranked according to their evaluated affinity for the selected domain/s. In the ‘more sequences’ mode, a list of SWISS-PROT–TrEMBL accession codes can be submitted for prediction. In this case, the output shows the best ranking peptide for each one of the submitted sequences. Peptides scoring lower than a fixed threshold are not displayed.
Figure 2.

WW domains prediction form. The list of all the available WW domains is reported. For each WW domain, the name of the protein and domain range sequence are reported as in the Pfam multiple alignment. Links to the InterPro, Pfam and SMART pages dedicated to the WW domains are available. A further link is provided for submitting lists of SWISS-PROT–TrEMBL accession codes.
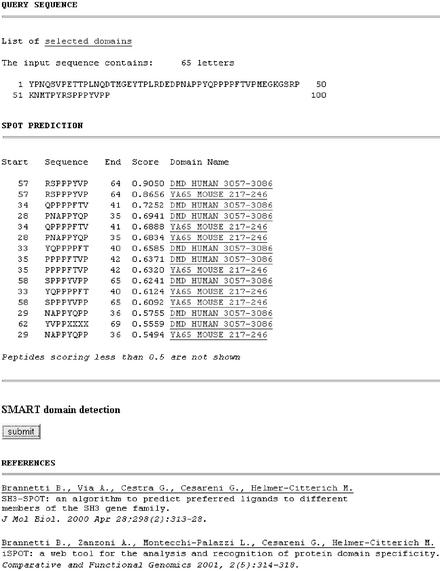
Figure 3.
Example of an output returned to the user. The WW domains are ranked according to their evaluated propensity to bind any peptides within the input sequence. For each eight-residue long peptide, the peptide sequence and position in the protein are listed. Then the score and the name of the WW domain used for the prediction are reported. The input sequence is reported as well, together with the number of all residues contained and, in case, the number of the non-supported characters (see the on-line documentation). A button is present for immediate submission of the query sequence to the SMART server.
WW DOMAINS VERSUS SWISS-PROT
For each available WW domain in the SPOT multiple alignment, potential binding partners found in the SWISS-PROT database (13) are reported. For each WW domain and query protein, only the highest-scoring peptide is reported. If the user wants to focus on a specific WW domain/SWISS-PROT sequence pair, the protein sequence in FASTA format can be retrieved (clicking on the available link) and then used as a query sequence in the ‘Protein sequence versus one or more WW domain sequences’ prediction form.
INTERACTION DATA
Experimentally derived interaction data have been used to build the WW domain database of residue–residue contact frequencies. These data, together with their references, are accessible in the third link available from the iSPOT query form (Fig. 1).
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by EEC project QLRI-CT-2000-00127 and by the Telethon multi-centre project GP0101Y01.
REFERENCES
- 1.Pawson T. and Nash,P. (2000) Protein–protein interactions define specificity in signal transduction. Genes Dev., 14, 1027–1047. [PubMed] [Google Scholar]
- 2.Gribskov M., McLachlan,A.D. and Eisenberg,D. (1987) Profile analysis: detection of distantly related proteins. Proc. Natl Acad. Sci. USA, 84, 4355–4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gribskov M. and Veretnik,S. (1996) Identification of sequence pattern with profile analysis. Methods Enzymol., 266, 198–212. [DOI] [PubMed] [Google Scholar]
- 4.Weng S., Dong,Q., Balakrishnan,R., Christie,K., Costanzo,M., Dolinski,K., Dwight,S.S., Engel,S., Fisk,D.G., Hong,E. et al. (2003) Saccharomyces Genome Database (SGD) provides biochemical and structural information for budding yeast proteins. Nucleic Acids Res., 31, 216–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gattiker A., Gasteiger,E. and Bairoch,A. (2002) ScanProsite: a reference implementation of a PROSITE scanning tool. Appl. Bioinf., 1, 107–108. [PubMed] [Google Scholar]
- 6.Rice P., Longden,I. and Bleasby,A. (2000) EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet., 16, 276–277. [DOI] [PubMed] [Google Scholar]
- 7.Smith G.R. and Sternberg,M.J. (2002) Prediction of protein-protein interactions by docking methods (2002). Curr. Opin. Struct. Biol., 12, 28–35. [DOI] [PubMed] [Google Scholar]
- 8.Sternberg M.J.E. Protein structure prediction—principles and approaches. In Sternberg,M.J.E. (ed.) Protein Structure Prediction, a Practical Approach, IRL Press, Oxford University pp. 1–30. [Google Scholar]
- 9.Wollacott A.M. and Desjarlais,J.R. (2001) Virtual interaction profiles of proteins. J. Mol. Biol., 313, 317–342. [DOI] [PubMed] [Google Scholar]
- 10.Brannetti B., Via,A., Cestra,G., Cesareni,G. and Helmer-Citterich,M. (2000) SH3-SPOT: an algorithm to predict preferred ligands to different members of the SH3 gene family. J. Mol. Biol., 28, 313–328. [DOI] [PubMed] [Google Scholar]
- 11.Brannetti B., Zanzoni,A., Montecchi-Palazzi,L., Cesareni,G. and Helmer-Citterich,M. (2001) iSPOT: a web tool for the analysis and recognition of protein domain specificity. A presentation for the ESF workshop ‘Proteomics: Focus on Protein Interactions’. Comp. Func. Genomics, 2, 314–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M.C., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O'Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]