Abstract
To date, assembling comprehensive annotation information for all probe sets of any Affymetrix microarrays remains a time-consuming, error-prone and challenging task. ChipInfo is designed for retrieving annotation information from online databases such as NetAffx and Gene Ontology and organizing such information into easily interpretable tabular format outputs. As companion software to dChip and GoSurfer, ChipInfo enables users to independently update the information resource files of these software packages. It also has functions for computing related summary statistics of probe sets and Gene Ontology terms. ChipInfo is available at http://biosun1.harvard.edu/complab/chipinfo/.
INTRODUCTION
As high-density oligonucleotide microarray technology becomes widely used, many microarray data analysis and visualization software packages have been implemented, such as dChip (1,2) and GoSurfer (http://biosun1.harvard.edu/complab/gosurfer/). dChip gives improved estimation of gene expression levels from modeling probe-level data and contains high-level analysis functions such as clustering and mapping genes to chromosome. GoSurfer visualizes Gene Ontology (GO) information of one to two gene lists and identifies GO categories enriched in a gene list. With the help of such software, microarray analysis has become manageable by biological and medical researchers at large. However, the developers of these software packages have been preparing and updating the accompanying information files, such as gene annotation files and the GO structure file. Constructing these files often requires the developers to download, parse and merge several online database files. This is a time-consuming process and is error-prone. More importantly, since most databases are updated constantly, software developers may not update the information files in a timely manner and the users suffer from not being able to utilize the latest information in their analyses.
To address these issues, we have developed ChipInfo (http://biosun1.harvard.edu/complab/chipinfo/), a software allowing researchers to download NetAffx (3) and Gene Ontology (4) database files, and reorganize, synthesize and output information files for dChip and GoSurfer. This software also does calculations to address interesting questions such as: what are the probes that are targeting homologous genes on different types of Affymetrix microarrays? Given an Affymetrix microarray, how many GO terms is every probe set associated with? What are the parent GO terms of a GO term?
SOFTWARE
ChipInfo is a single executable program downloadable from the web. It runs on Windows 98/2000/NT/XP. Supplementary materials including a manual and some application examples are available at http://biosun1.harvard.edu/complab/chipinfo/.
Accessing online databases
ChipInfo uses the NetAffx and the Gene Ontology databases. By choosing either ‘NetAffx’ or ‘Gene Ontology’ from the menu ‘Annotation→Accessing Web Databases’, ChipInfo will pop up a dialog box for users to choose from ‘Automatic Access’ or ‘Interactive Access’. The ‘Automatic Access’ function will check whether the target database has been updated since the last time the user accessed it, and if yes, link to the target database and download the needed files to replace the older versions of these files on the local machine. The ‘Interactive Access’ function starts Internet Explorer to browse the web page of the target databases. Meanwhile, instructions are shown in the software to guide users through querying the databases and downloading files. In addition, ChipInfo reports the time of the user's latest access to the target database and the jobs the user has completed to help the user to decide whether to update the local data files. The downloaded data files are then used as the input data for ChipInfo, as described in the next section.
Constructing Gene Information File
A Gene Information File is a tabular text file, annotating all the probe sets of a given Affymetrix array type. Each line of it contains the annotation information of a probe set, including GenBank ID, LocusLink ID (5), gene name, GO terms, protein domain terms, signaling pathways, cytoband and Affymetrix descriptions. Table 1 shows what a Gene Information File looks like.
Table 1. Part of a Gene Information File constructed by ChipInfo for Affymetrix U74av2 array. Due to space limitation, a large part of the annotations are replaced manually by ‘…’.
| Probe set | GenBank | LocusLink | Name | Gene Ontology | Protein domain | Pathway | Chrom. Desc. |
|---|---|---|---|---|---|---|---|
| 100001_at | M18228 | 12502 | CD3 antigen, gamma polypeptide | |7166|7165|… | |3598|3110| | M18228 | |9| … |
| 100002_at | X70393 | 16426 | inter-alpha trypsin inhibitor, heavy chain 3 | |4867|4866|… | |1117|2035| | |14| … | |
| 100016_at | Z12604 | 17385 | Matrix metalloproteinase11 | |6805|9410|… | |6026|1818|… | |Matrix Metalloproteinases| | |10| … |
| 100017_at | U68267 | 53311 | myosin binding protein H | |7517|9887|… | |3598|3961|… | |Cytoskeletal Pathway|… | |1| … |
Gene Information Files are useful input files to both dChip and GoSurfer. In the past we needed to download large data files from the UniGene (6) database, parse them and generate a mapping from LocusLink ID to Unigene ID. The same procedure was taken for the LocusLink database to obtain the GO and protein domain information for each LocusLink ID. Finally these mappings had to be linked altogether to produce the Gene Information Files. Several recent large-scale and systematic efforts including Resourcerer (7), AnnBuilder (8) and NetAffx have been made to generate such mappings and produce online databases. ChipInfo downloads the NetAffx data that are relevant to a particular array type and processes them to generate Gene Information Files directly usable by dChip or GoSurfer. Figure 1 shows the ‘Annotation/Gene Information File’ function for generating Gene Information Files.
Figure 1.
Making Gene Information Files with ChipInfo. When the ‘Annotation→Gene Information File’ menu is clicked, a ‘Make gene information file’ dialog shows up. Designate a NetAffx file and a Gene Ontology file in the dialog as ChipInfo's inputs. The preparation method for these two files is described in the Accessing Web Databases section. The user can change the name and format of the output file.
Besides the NetAffx database, ChipInfo also uses the Gene Ontology (4) files ( process.ontology, function.ontology and component.ontology) to enrich the GO annotation for probe sets. Because of the structural relationships among GO terms, every probe set associated with one GO term is also associated with all its ancestor GO terms. However, online databases such as LocusLink only contains the lowest level GO terms for a gene, but not their ancestor GO terms. ChipInfo reads in these lowest level GO terms for a probe set and traces the GO structure to retrieve all their ancestor terms, thus explicitly linking every probe set to their complete GO annotation in the Gene Information Files.
ChipInfo also allows users to set the maximum number of GO terms used in the Gene Information Files. This function is provided for users to eliminate the GO terms that associate with very few probe sets. Usually a large number of GO terms are associated with at least one probe set on an Affymetrix array, but most GO terms are each associated with only a very small number of probe sets. If a maximum number of GO terms is set by the user, ChipInfo will rank all GO terms according the number of probe sets they associate with and only keep the given number of top ranking GO terms.
Constructing Gene Ontology structure file
Since Gene Ontology files (process.ontology, function.ontology and component.ontology) are organized in a pairwise ‘parent–child’ manner, they cannot be directly used in GoSurfer. ChipInfo downloads, parses and reorganizes GO files into a GO structure file usable in GoSurfer. The GO structure file contains the information of GO IDs, a path code for every GO path (a sorted collection of a GO term and all its ancestor terms) and the mapping between the GO terms and path codes. Such information explicitly gives the position that every GO term belongs to in the GO graph and greatly eases the time-consuming procedure of positioning of GO terms in GO visualizing software packages. GO structure file is generated at the ‘Annotation→Gene Information File’ dialog. Figure 2 illustrates how the path code works. Table 2 shows a partial Gene Ontology structure file.
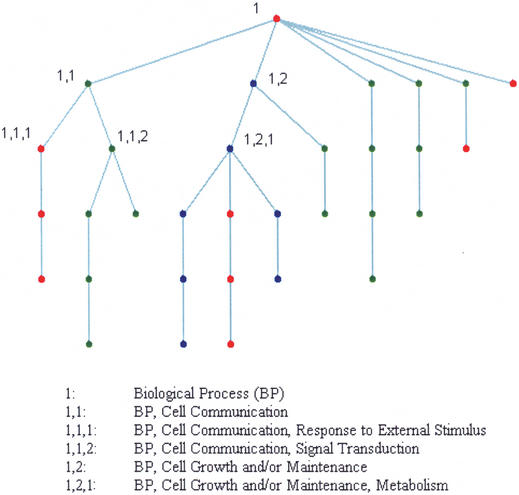
Figure 2.
Example of GO path code. A GO term ‘Cell Growth and/or Maintenance’ is denoted by path code ‘1,2’ and its position in the GO graph is uniquely determined. This idea was originated by Ming-Chih Kao and first used by Storch et al. (11).
Table 2. Partial GO structure file.
| GO ID | Path code | Real path |
|---|---|---|
| 3674 | 2 | Molecular Function (MF) |
| 15643 | 2,1 | MF, anti-toxin |
| 15644 | 2,1,1 | MF, anti-toxin, lipoprotein anti-toxin |
| 5488 | 2,6 | MF, binding |
| 16597 | 2,6,1 | MF, binding, amino acid binding |
| 16595 | 2,6,1,1 | MF, binding, amino acid binding, glutamate binding |
| 16594 | 2,6,1,2 | MF, binding, amino acid binding, glycine binding |
Calculating ‘array background’
After an array type is chosen by the user, ChipInfo calculates the number of probe sets associated with every GO term and the number of GO terms associated with every probe set. These ‘array background’ results can be exported as tabular files. They enable researchers to compare a subset of genes to all the genes on an array to identify significantly enriched GO terms in this subset (9,10) (more information about this comparison method is available at http://biosun1.harvard.edu/complab/gosurfer/). Users can check the ‘Calculate array background’ option in the ‘Make Gene Information File’ dialog to perform this function.
Retrieving probe sets for homologous/orthologous genes
Researchers are often interested in how to obtain all the identical, homologous or orthologous probe set pairs among different array types. For example, a mouse or rat model for a human disease is constructed and array data are obtained from both human samples and animal models. Consistent gene expression changes across human and animal models may cross-validate the results. The key to this analysis is to obtain the mapping of the probe sets targeting homologous or orthologous genes. To our knowledge, so far researchers have to do manual work to get such information. NetAffx files contain homologous/orthologous probe set information among human, mouse and rat arrays. Table 3 shows a partial NetAffx file where homologous/orthologous probe set information is displayed. ChipInfo extract such information to make ‘common probe set’ files containing homologous/orthologous probe set pairs between two user-defined array types. Please be aware that as a data extraction tool, ChipInfo can only be as accurate as the underlining database. To use the homologous/orthologous probe set retrieval feature, users can select the ‘Annotation→Common Probe Set File’ menu and choose two array types in the dialog. If the two chosen array types are for different species, the probe set pairs for homologous or orthologous genes will be exported. If the two array types are for the same species, the probe sets targeting the same gene will be exported. It is possible that more than one probe sets in one array target the same gene or one gene of one species has several homologous or orthologous genes in the other species. In this case, all possible probe set pairs will be exported. The exported Common Probeset File can be used in dChip to combine the data across array types or species for analysis.
Table 3. Partial NetAffx file. This is a small fraction of the NetAffx file containing information for the human HG-U95Av2 array. Mouse and rat homologous/orthologous probe sets are listed under the ‘Orthologs/Homologs’ entry for this HG-U95Av2 probe set.
| ID | 1000_at HG-U95Av2 |
| Title | mitogen-activated protein kinase 3 |
| Sequence ID | X60188mRNA |
| … | |
| Orthologs/Homologs | |
| Rat | M61177_s_at RG-U34A; mitogen activated protein kinase 3; Curated Ortholog |
| M61177_s_at RN-U34; mitogen activated protein kinase 3; Curated Ortholog | |
| rc_AI235753_at RG-U34C; mitogen activated protein kinase 3; Curated Ortholog | |
| M61177_s_at RT-U34; mitogen activated protein kinase 3; Curated Ortholog | |
| 1370898_a_at RAE230A; mitogen activated protein kinase 3; Curated Ortholog | |
| Mouse | Msa.2276.0_s_at Mu11KsubB; mitogen activated protein kinase 3; Curated Ortholog |
| 101834_at MG-U74Av2; mitogen activated protein kinase 3; Curated Ortholog |
DISCUSSION
Constructing and updating biological information for microarray analysis becomes more important than ever due to fast updates and changes in public databases (for example, NetAffx is updated quarterly). ChipInfo links public databases and microarray analysis software and provides users the ability to quickly utilize the latest information in their microarray analyses.
Even though ChipInfo relies heavily on NetAffx, there are several important differences between using ChipInfo and using NetAffx's web interface. While NetAffx can only be queried interactively for one or a few genes at a time, ChipInfo provides information files to be used in dChip (V1.3) and GoSurfer (V1.0) for large-scale microarray analysis. In addition, ChipInfo traces the GO structure to obtain the full GO annotations of a probe set, based on the lowest level GO term annotations provided by LocusLink or NetAffx. Moreover, ChipInfo output files are in Excel-like tabular format and thus more interpretable and users may process them further to get certain information or produce information resource files for other applications.
ChipInfo is an open-source project and we welcome joint efforts to continue the development. Some specific features that we are considering to add in the future version are: column-wise customization of Gene Information File, so users can decide what information columns to be included; constructing information files for other software; retrieving information from other databases such as LocusLink (5), ‘Mouse Genome Informatics’ database (http://www.informatics.jax.org/) and ‘UCSC Genome Bioinformatics’ database (http://genome.ucsc.edu/), so that we may get annotation information not included in NetAffx and make annotation files for cDNA or protein microarrays.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Robert Gentleman, Thomas Seidl, Nik Brown, Mike Thompson and two reviewers for helpful discussions. This work is supported by NIH grants 1R01HG02341 and P20 CA96470.
NOTE ADDED IN PROOF
In March 2003, NetAffx made Annotation CSV files available online. These files have a tabular format and contain information similar to that in Table 3, but are easier to download. Accordingly, we have updated ChipInfo to read such new file formats.
REFERENCES
- 1.Li C. and Wong,W.H. (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl Acad. Sci. USA, 98, 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li C. and Wong,W.H. (2001) Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol., 2, research0032.1–0032.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu G., Loraine,A.E., Shigeta,R., Cline,M., Cheng,J., Valmeekam,V., Sun,S., Kulp,D. and Siani-Rose,M.A. (2003) NetAffx: Affymetrix probesets and annotations. Nucleic Acids Res., 31, 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T. et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuler G.D. (1997) Pieces of the puzzle: expressed sequence tags and the catalog of human genes. J. Mol. Med., 75, 694–698. [DOI] [PubMed] [Google Scholar]
- 7.Tsai J., Sultana,R., Lee,Y., Pertea,G., Karamycheva,S., Antonescu,V., Cho,J., Parvizi,B., Cheung,F. and Quackenbush,J. (2001) RESOURCERER: a database for annotating and linking microarray resources within and across species. Genome Biol., 2, software0002.1–0002.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J., Carey,V. and Gentleman,R. (2003) An extensible application for assembling annotation for genomic data. Bioinformatics, 19, 155–156. [DOI] [PubMed] [Google Scholar]
- 9.Cho R.J., Huang,M., Campbell,M.J., Dong,H., Steinmetz,L., Sapinoso,L., Hampton,G., Elledge,S.J., Davis,R.W. and Lockhart,D.J. (2001) Transcriptional regulation and function during the human cell cycle. Nature Genet., 27, 48–54. [DOI] [PubMed] [Google Scholar]
- 10.Tavazoie S., Hughes,J.D., Campbell,M.J., Cho,R.J. and Church,G.M. (1999) Systematic determination of genetic network architecture. Nature Genet., 22, 281–285. [DOI] [PubMed] [Google Scholar]
- 11.Storch K.F., Lipan,O., Leykin,I., Viswanathan,N., Davis,F.C., Wong,W.H. and Weitz,C.J. (2002) Extensive and divergent circadian gene expression in liver and heart. Nature, 417, 78–83. [DOI] [PubMed] [Google Scholar]