Abstract
The Vienna RNA secondary structure server provides a web interface to the most frequently used functions of the Vienna RNA software package for the analysis of RNA secondary structures. It currently offers prediction of secondary structure from a single sequence, prediction of the consensus secondary structure for a set of aligned sequences and the design of sequences that will fold into a predefined structure. All three services can be accessed via the Vienna RNA web server at http://rna.tbi.univie.ac.at/.
INTRODUCTION
Biomolecules exhibit a close interplay between structure and function. Therefore the growing number of RNA molecules with complex functions, beyond that of encoding proteins, has brought increased demand for RNA structure prediction methods. While prediction of tertiary structure is usually infeasible, the area of RNA secondary structures is an example where computational methods have been highly successful.
The first practical dynamic programming algorithms to predict the optimal secondary structure of an RNA sequence date back over 20 years (1). Since then they have been extended to allow prediction of suboptimal structures (2,3) and thermodynamic ensembles (4), which allow to assign a confidence level or ‘well definedness’ to the predictions (5).
Recently, several methods have addressed the problem of predicting a consensus structure for a group of related RNA sequences (6–11). Such conserved structures are of particular interest, since conservation of structure in spite of sequence variation implies that the structure must be functionally important. By enhancing energy rules with sequence covariation these methods also obtain much better prediction accuracies.
The Vienna RNA package (12) is a free software package that implements a variety of algorithms for the prediction and analysis of RNA secondary structures. The package is, however, strongly geared toward Unix command-line users and programmers. For the less computer savvy, or occasional user, it provides neither a point-and-click graphical user interface nor even pre-compiled binaries.
The Vienna RNA web site tries to address these shortcomings by offering access to the most popular features via an easy to use web interface. It consists of three CGI scripts equivalent to the RNAfold, RNAalifold and RNAinverse command line programs, respectively. While the servers have to limit request sizes for performance reasons, they return for each request an equivalent command line invocation. This makes it easier for users to make the transition to locally installed software, should their requirements exceed the limits of the web service.
THE RNAfold SERVER
Of the three services, the RNAfold server provides both the most basic and most widely used function. Input consists of a single sequence that has to be typed or pasted into a text field of the input form.
In the simplest case, the server predicts only the minimum free energy (mfe) structure of a single sequence using the classic algorithm of Zuker and Stiegler (1). In addition to mfe folding the server can calculate equilibrium base pairing probabilities via John McCaskill's partition function algorithm (4).
By default the RNA energy parameters of the Turner group (13) are used, but single stranded DNA sequences can be handled as well, by selecting the DNA parameter set provided by John SantaLucia (14).
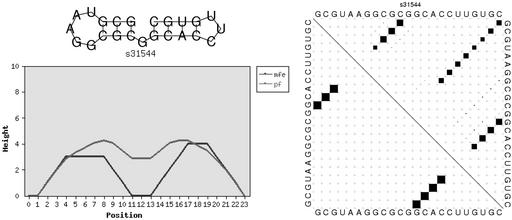
The fold server output consists of a static html page presenting the predicted mfe structure as a string in bracket notation and links to the plots generated for visualization. Three types of plots can be produced. Firstly, the predicted mfe structure is plotted as a conventional secondary structure graph using the naview layout method (15). The pair probabilities can be visualized in a so-called ‘dot plot’: on a square grid of n×n we draw for each possible pair (i, j) a box with area proportional to its probability. Finally, we produce a mountain plot depicting both the predicted mfe and pair probabilities. A mountain plot is an xy-graph that plots the number of base pairs enclosing a sequence position (for pair probabilities the average number of enclosing pairs). See Figure 1 for examples of all three representations.
Figure 1.
Three representations for secondary structure and structure ensembles as produced by the RNAfold service: structure graph, mountain plot and dot plot. As can be seen clearly in the dot plot, the sequence has two almost equally good but very dissimilar foldings.
Secondary structure drawing and dot plots are always produced in Postscript format. Postscript is used not only because it gives the highest print quality, but also because it allows the actual data to be embedded in the file, e.g. all pair probabilities are contained in the dot plot in an easy to parse format. On the other hand, Postscript files cannot be used for inline images on web pages and require additional software for viewing (e.g. gsview, http://www.ghostscript.com/).
A suitable alternative is the new standard for Scalable Vector Graphics, SVG (http://www.w3.org/Graphics/SVG). Users with SVG enabled browsers (typically through the use of Adobe's SVG plugin, http://www.adobe.com/svg/) can request structure drawings in SVG, which allows some interactivity such as toggling annotation. Currently the server accepts sequences up to a maximum length of 4000 nt, sequences up to 300 nt will be processed immediately while longer jobs are submitted to a batch queue, in which case the user is notified by email after completion.
THE Alifold SERVER
The Alifold service predicts the consensus secondary structure for a set of aligned RNA or DNA sequences by using modified dynamic programming algorithms that add a covariance term to the standard energy model (11), again it supports prediction of mfe structures and pair probabilities. Usage is almost identical to that of the RNAfold service. Instead of typing an input sequence, a precomputed sequence alignment is uploaded via the input form. Currently, only alignments in Clustal format are accepted. The server restricts both the size of the upload and the length of the alignment, current limits being 10 Kb and 2000 nt, respectively.
Results are again visualized in Postscript plots that are enhanced by information on sequence variation. In the structure drawings mutations supporting the predicted structure are marked by circles, in the dot plots and mountain plots, color is used to indicate the number of different pair types. Examples and detailed explanation of these representations can be found on the online help page (http://www.tbi.univie.ac.at/~ivo/RNA/alifoldcgi.html).
THE INVERSE FOLD SERVER
Finding sequences that fold into a predefined structure is the inverse of structure prediction problem. Often it is useful to design such sequences, e.g. in order to experimentally test an hypothesis about functional structures. While this is often done manually for very short sequences, it quickly becomes tedious and error prone.
Our inverse folding service treats sequence design as an optimization problem in sequence space that is solved heuristically (12). There are again two variants based on mfe and partition function folding. In the first case we minimize the dissimilarity between the predicted mfe structure and the desired target structure. In the second case we optimize the frequency of the target structure in the thermodynamic ensemble. While the mfe optimization typically yields sequences that are marginally stable, i.e. have many alternative foldings, optimization via the partition function produces sequences with a very strong preference for the target structure.
Input consists simply of the desired structure in bracket notation. The maximum structure length is currently 100 nt. The time needed for the search varies widely depending on the ubiquity of the target structure. Most valid secondary structure strings never occur as mfe structure of some sequence (i.e. many sequence design problems have no solution), while some others are extremely common (for example see 16). Conversely, the number of search steps performed by the algorithm is a good indicator for the frequency of a structure in sequence space.
FUTURE PLANS
The Vienna RNA secondary structure server presented here provides only basic access to a subset of the functions in the Vienna RNA software package. Nevertheless they provide a convenient interface for users that need RNA structure prediction only occasionally and a shallow learning curve for those new to the field.
Work is underway to further improve the visualization of the results, e.g. by producing structure drawings annotated with various measures of well-definedness. As SVG enabled browsers become more widespread, a combination of SVG graphics and client side javascript should allow users to explore the predicted structures interactively.
The output web page produced by the server is designed for the interactive user and thus is not ideal for automatic parsing and further processing of the results. To facilitate such interoperation with other programs and web services we plan to offer input and output in a standardized data exchange format. A promising candidate for this is the recently proposed RNAML format (17), an XML based language for the storage of information on RNA sequence and structure.
While the server currently runs on a somewhat dated dual Pentium II 450 MHz machine, the use of a batch queuing system allows jobs to be distributed to other machines should that become necessary.
Acknowledgments
ACKNOWLEDGEMENTS
This work is supported by the Austrian Fonds zur Förderung der Wissenschaftlichen Forschung, Projects FWF 15893 and P-13545-MAT.
REFERENCES
- 1.Zuker M. and Stiegler,P. (1981) Optimal computer folding of larger RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res., 9, 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zuker M. (1989) The use of dynamic programming algorithms in RNA secondary structure prediction. In Waterman,M.S. (ed.), Mathematical Methods for DNA Sequences, CRC Press, Boca Raton, FL, pp. 159–184. [Google Scholar]
- 3.Wuchty S., Fontana,W., Hofacker,I.L. and Schuster,P. (1999) Complete suboptimal folding of RNA and the stability of secondary structures. Biopolymers, 49, 145–165. [DOI] [PubMed] [Google Scholar]
- 4.McCaskill J.S. (1990) The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers, 29, 1105–1119. [DOI] [PubMed] [Google Scholar]
- 5.Zuker M. and Jacobson,A.B. (1995) ‘Well-determined’ regions in RNA secondary structure prediction: analysis of small subunit ribosomal RNA. Nucleic Acids Res., 23, 2791–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorodkin J., Heyer,L.J. and Stormo,G.D. (1997) Finding the most significant common sequence and structure motifs in a set of RNA sequences. Nucleic Acids Res., 25, 3724–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hofacker I.L., Fekete,M., Flamm,C., Huynen,M.A., Rauscher,S., Stolorz,P.E. and Stadler,P.F. (1998) Automatic detection of conserved RNA structure elements in complete RNA virus genomes. Nucleic Acids Res., 26, 3825–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lück R., Graf,S. and Steger,G. (1999) Construct: a tool for thermodynamic controlled prediction of conserved secondary structure. Nucleic Acids Res., 27, 4208–4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juan V. and Wilson,C. (1999) RNA secondary structure prediction based on free energy and phylogenetic analysis. J. Mol. Biol., 289, 935–947. [DOI] [PubMed] [Google Scholar]
- 10.Knudsen B. and Hein,J. (1999) RNA secondary structure prediction using stochastic context-free grammars and evolutionary history. Bioinformatics, 15, 446–454. [DOI] [PubMed] [Google Scholar]
- 11.Hofacker I.L., Fekete,M. and Stadler,P.F. (2002) Secondary structure prediction for aligned RNA sequences. J. Mol. Biol., 319, 1059–1066. [DOI] [PubMed] [Google Scholar]
- 12.Hofacker I.L., Fontana,W., Stadler,P.F., Bonhoeffer,S., Tacker,M. and Schuster,P. (1994) Fast folding and comparison of RNA secondary structures. Monatsh. Chem., 125, 167–188. [Google Scholar]
- 13.Mathews D., Sabina,J., Zucker,M. and Turner,H. (1999) Expanded sequence dependence of thermodynamic parameters provides robust prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 14.SantaLucia J. Jr. (1998) A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA, 95, 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruccoleri R.E. and Heinrich,G. (1988) An improved algorithm for nucleic acid secondary structure display. CABIOS, 4, 167–173. [DOI] [PubMed] [Google Scholar]
- 16.Schuster P., Fontana,W., Stadler,P.F. and Hofacker,I.L. (1994) From sequences to shapes and back: a case study in RNA secondary structures. Proc. Royal Society London B, 255, 279–284. [DOI] [PubMed] [Google Scholar]
- 17.Waugh A., Gendron,P., Altman,R., Brown,J., Case,D., Gautheret,D., Harvey,S., Leontis,N., Westbrook,J., Westhof,E. et al. (2002) RNAML: a standard syntax for exchanging RNA information. RNA, 8, 707–717. [DOI] [PMC free article] [PubMed] [Google Scholar]