Abstract
FIE2 (5′ end Information Extraction v2) is a web-based program for easy identification and extraction of nucleotide sequence around the start of genes (promoter region) and their translation initiation site (TIS). Using information provided by the National Center for Biotechnology Information's (NCBI's) LocusLink, FIE2 identifies the 5′-most end of a gene on its respective chromosome based on alignment of a selected set of mRNAs representative of the gene. FIE2 then uses currently available human genome sequence information to extract the desired sequences. The accuracy of the information extracted is therefore limited by the accuracy and completeness of the sequence annotation and sequence alignment provided by LocusLink. In addition, multiple TIS positions are also occasionally presented, for example, as a result of multiple alignments of transcript variants. One of the key criteria of FIE2 is that it should extract only the correct information or attempt no extraction at all. To date, the authors are not aware of any publicly available web-based tool that uses the human genomic sequence to extract pertinent promoter- and TIS-region information in this fashion. FIE2 is freely available at http://sdmc.lit.org.sg/FIE2.0.
INTRODUCTION
The study of gene expression has, of late, extended its tentacles from the wet laboratories to the computer-intensive Bioinformatics laboratories. Regulation of gene expression is mediated mainly through the promoter. Promoters are stretches of DNA sequences, generally located upstream of, and overlapping, the transcription start site (TSS) of genes. There is an abundance of mRNA/cDNA sequence information from public databases, such as GenBank, which are available to molecular biology researchers and bioinformaticians for study. However, therein lies a key problem: while information is plentiful and readily available, the information may also be disparate and incomplete. For example, mRNA sequences for a particular gene may be of varying length because different laboratories who have attempted to clone the gene may have achieved this with varying degrees of success; therefore some of the mRNA sequences entered into GenBank may be 5′-incomplete. The solution is to try and find a way to filter out as much valuable information as possible from these public databases such that we may use all these sequence information to study the characteristics of the gene start region in order to gain a better insight into gene expression.
FIE2 is a specialized program for the extraction of genomic DNA sequences around the start (promoter region) and translation initiation site (TIS) of a gene. The start and TIS positions of the gene are determined from the alignment of a set of mRNAs representative of the gene of interest on the human working draft genomic sequence, as given by LocusLink's Evidence Viewer (1–3). As a result of multiple alignments of mRNAs on the genomic sequence, multiple start positions are usually given for a gene. The 5′-most SOE1 (‘start of exon 1’) position identified by FIE2 thus represents the 5′-most position of the alignments of the representative mRNA transcript(s) on the genomic contig. Currently, there are only two other programs, Promoter Extraction from GenBank (PEG) (4) and EZ-Retrieve (5), which although are similar in their goal to FIE2, differ from FIE2 in functionality and methodology: primarily PEG draws its extraction from GenBank's mRNA records instead of the human working draft genomic sequences while EZ-Retrieve uses the Abstract-Syntax-Notation-One (ASN.1) files to get an approximate position of the gene's start which is not always supported by gene transcripts. Furthermore, both PEG and EZ-Retrieve cannot extract sequences around the TIS and PEG is also not accessible from the web.
The importance of the sequences extracted by FIE2 also lies in its usefulness for follow-up experiments in the laboratory in current research efforts to understand the transcriptional machinery. In addition, the sequences can also be used to compile datasets for training and testing gene finding/prediction systems, such as Dragon Promoter Finder (6,7) and Dragon Gene Start Finder (see http://sdmc.lit.org.sg/promoter).
PROGRAM DESCRIPTION
FIE2 can be accessed at the given URL address. The web interface for FIE2 is fairly intuitive. Input to FIE2 can either be a gene or protein name or LocusID (for additional query options, please refer to LocusLink's help page: http://www.ncbi.nlm.nih.gov/LocusLink). Users must also input the length of sequence upstream and downstream of the start of the gene (which is abbreviated, SOE1 [‘start of exon 1’], by FIE2) that they wish to extract. The user is encouraged to be as specific as possible when submitting a query to FIE2; for example, where possible, users should use the option to search by LocusID. If a general query (e.g. actin) is submitted to FIE2, this is sent to LocusLink which then returns a list of links to genes which match the query. The user must choose the appropriate link for the gene of his interest. The user's request is again submitted to LocusLink which then returns an information page which is processed by FIE2. Among the information that FIE2 gathers from LocusLink's information page is the availability of an Evidence Viewer (EV) page. For human genes, LocusLink attempts to align a set of published sequences representative of a gene on its respective chromosomal sequence (genomic contig) in its EV page (Fig. 1A). The number and specific instances of accession numbers (gene records) used in the alignment depend on whether the gene has a provisional or reviewed reference sequence (RefSeq, http://www.ncbi.nlm.nih.gov/LocusLink/refseq.html) record, or no RefSeq record at all. If no EV page is available, FIE2 returns a ‘No Evidence Viewer found’ page. If the initial query submitted to FIE2 is very general (e.g. actin), LocusLink would return a fairly long list of gene links. Unfortunately, there is no way for FIE2 to determine if the gene links contain an EV page without making several hits to the LocusLink server to gather the LocusLink information page for each and every one of those genes on the list (this is an undesirable practice since it places an unnecessary burden on the LocusLink server). One may convert accession numbers to LocusID values using the daily updated file that is available from ftp://ncbi.nlm.nih.gov/refseq/LocusLink/loc2acc.
Figure 1.
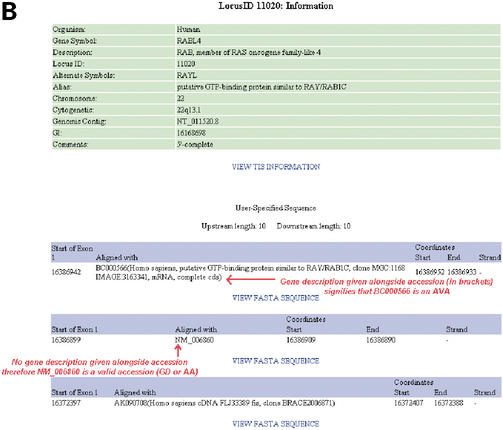
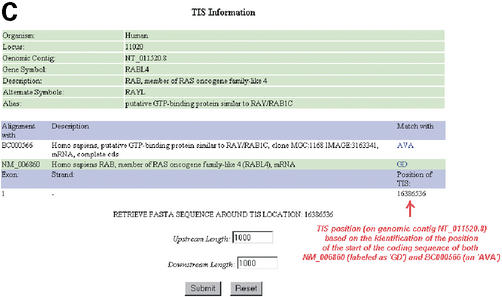
(A) An example of LocusLink's EV page. In this EV page, the alignment of representative mRNA sequences for the gene RABL4 (LocusID: 11020) is shown. NM_006860 is recognized as a valid accession for the gene RABL4 through its exact match of it gene name/description and therefore classed as ‘GD’ (a valid accession) by FIE2. BC000566 (an IMAGE Consortium cDNA clone) is recognized as a possible mRNA sequence representative of the gene RABL4 because of its high degree of identity to the known RefSeq sequence for RABL4 (NM_006860). BC000566 is therefore classed as an associated valid accession or ‘AVA’ by FIE2. As can be seen here, the start of the coding sequence is found in ‘Exon 1’ of the genomic region present on the EV page. All labels in red were added by the authors for clarity of understanding. (B) An example of FIE2's SOE1 Information page. Here, we see that three SOE1 positions are given for RABL4 based on the alignment of two AVAs (BC000566 and AK090708) and a valid accession (NM_006860). The accession numbers for AVAs are followed by their gene description whereas no such description is given for the valid accession, NM_006860. The choice is left to the user to determine whether he wishes to use the sequence extracted based on the identified AVAs or valid accession. A ‘View FASTA Sequence’ hyperlink is provided for each identified SOE1 position. The 5′-most SOE1 position is always given first on the page followed by the next 5′-most SOE1 position identified. All labels in red were added by the authors for clarity of understanding. (C) An example of FIE2's TIS Information page. As can be seen here, the identified TIS position for RABL4 is supported by coding sequences of both NM_006860 (a valid accession, denoted as ‘GD’ on the page) and BC000566 (an AVA). All labels in red were added by the authors for clarity of understanding.
It is possible that the LocusLink EV page presents more than one gene along a genomic region of the contig. FIE2 attempts to recognize all relevant (valid) accessions (mRNA sequences) by gene name or symbol or other aliases among the accessions presented in the sequence alignment on the EV page. The valid accessions are abbreviated ‘GD’ or ‘AA’ by FIE2 depending on whether the accession was identified as valid based on a match of its gene name/description/symbol or an alias/alternative symbol, respectively (Fig. 1C). Based on the sequence alignment given on the EV page, if an accession has a high sequence identity with the 5′ end exon of an already identified valid accession (but was not previously recognized as a valid accession by its gene name or symbol), then FIE2 labels this as an ‘AVA’ (associated valid accession) (Fig. 1A and C). The description of the AVA is given alongside its accession on the result page bearing the SOE1 position information (no descriptions are given for valid accessions, GD and AA) (Fig. 1B). The SOE1 positions, based on the alignment of all valid accessions and AVAs on the genomic contig, are presented to the user for his analysis. The FASTA sequence (with the user-specified length) around all the SOE1 positions identified by FIE2 can be retrieved through their respective ‘View FASTA Sequence’ hyperlink (Fig. 1B). The 5′-most SOE1 position identified by FIE2 thus represents the 5′-most position of the alignments of the representative mRNA transcript(s) on the genomic contig.
Along with the DNA sequence alignment on the genomic contig, the EV page also presents an alignment of the coding sequence of each accession alongside its DNA sequence. For each valid accession, FIE2 locates the position of the TIS on the genomic contig by identifying the position of the start of its coding sequence (Fig. 1A). If the TIS is found to be in ‘Exon 1’ of the genomic region presented on the EV page then the SOE1 position for each valid accession or AVA is identified individually based on the position of 5′-most end of the aligned mRNA sequence on the genomic contig. For example, in the case of RABL4, we can see that the start of the coding sequence is in ‘Exon 1’ (Fig. 1A) and the SOE1 positions based on the 5′-most position of NM_006860 and BC000566 on the contig are identified by FIE2 and displayed accordingly, as shown in Figure 1B. If, however, the TIS position does not meet the above criteria, then in addition to identifying the SOE1 position for each valid accession on the genomic contig, a process is also initiated to determine if ‘Exon 1’ presented on the EV page is indeed the first exon of the queried gene. This is because in certain cases, where there is more than one gene in the genomic region presented on the EV page, the first exon presented in the sequence alignment might not represent the first exon of the queried gene. In such instances, the true first exon is identified by locating the exon containing the 5′-most position of all the aligned mRNAs of the queried gene. FIE2 then renumbers all exons on the EV page accordingly so as to reflect the true exonic-intronic partition of the gene sequence on the genomic contig. This step is crucial in determining the exon(s) that contains the SOE1 and TIS.
For FIE2, multiple SOE1 positions may be presented (as explained above) and likewise, multiple TIS positions may also be given. The given coding sequence for some of the valid accessions or AVAs may sometimes be predicted and therefore, the position of the TIS differs from those of other valid accessions used to align against the genomic contig. The coding sequence and thus, the TIS is sometimes predicted and not experimentally verified by the laboratory which cloned the cDNA sequence; however, such information/annotation is not provided by LocusLink. Therefore, FIE2 presents all TIS positions (predicted or otherwise) for the sake of completeness. In some cases, the presence of multiple TISs may also be due to the different initiation sites for different transcript variants.
Several scenarios may present themselves that might leave FIE2 to tag the given SOE1 position as ‘indeterminate’ (i.e. FIE2 cannot determine the SOE1 position). Four categories of ‘indeterminate’ exist in FIE2 and these are explained in detail on the program's web site (in the FAQ page at http://sdmc.lit.org.sg/FIE2.0/faq.html). FIE2 still retrieves the sequence upstream and downstream of the 5′-most position of the mRNA alignment on the genomic contig in these cases, but with the caveat that the SOE1 position is ‘indeterminate’. It is up to the user's discretion to determine whether or not to use the sequence provided.
FIE2 also determines the strand orientation of the gene on the genomic contig. If the locus is found on the complementary strand of the contig, FIE2 retrieves the user-specified sequence region and presents the FASTA sequence in its reverse complement. A similar process is carried out to retrieve the FASTA sequence around the TIS where available. The header for the FASTA sequence can be interpreted as follows: for example, for LocusID 10043, if the user chooses to extract 10 bp upstream and downstream of the SOE1 position, then the header would read ‘>gi|16168698:14992135-14992154 Homo sapiens chromosome 22 reference genomic contig| SOE1|14992145’. This mean that the SOE1 for this gene (in this case, based on the alignment of AK026576) is found in the genomic contig (GI [NCBI's sequence identifier]: 16168698) for chromosome 22 and the SOE1 position is 14 992 145 on this contig. The 20 nt long sequence requested by the user therefore stretches from position 14 992 135–14 992 154 on the contig.
The following additional information on the gene of interest is also provided:
The descriptive name of the gene.
Alternate symbols/aliases for the gene.
The chromosome on which this genetic locus is found.
The gene's cytogenetic position on the above chromosome.
The accession number for the genomic contig on which this locus is found.
The GI (GenBank's unquie identifier) number for the contig.
Examples of the output returned by FIE2 can be viewed from the Chromosome 22 test dataset table (http://sdmc.lit.org.sg/FIE2.0/test-dataset.html) by clicking on one of the LocusID given there.
RESULTS
Testing FIE2
Recently, an updated annotation for human chromosome 22 was released (8). Therefore, we decided to use chromosome 22 to benchmark FIE2. There are altogether 393 mRNA sequences of protein coding genes which are considered complete and mapped to the genomic sequence for human chromosome 22 (http://www.sanger.ac.uk/cgi-bin/hgp/chr22/display?Chr22.3.1b.coding_genes.gff). Although not all known genes from the current Sanger chromosome 22 annotation are yet included in LocusLink, we could still find 230 in LocusLink. For these 230 genes, FIE2 could determine the SOE1 position for 208 and the SOE1 position for the remaining 22 genes were either tagged as ‘indeterminate’ or given a tag that represented the fact that there was no EV page or an incomplete EV page for that particular LocusID.
Of the 208 genes whose SOE1 position could be determined by FIE2, 40 matched exactly the gene start positions annotated by Sanger, while 54 were, in fact, found to be upstream of the positions given by Sanger. The SOE1 position extracted by FIE2 for the remaining 112 genes were all found to be downstream of Sanger's annotated positions. Even so, the SOE1 positions of 74 of the remaining 112 genes were within 100 bp of the annotated position given by Sanger. While 80.8% (168 of 208 genes) of the extracted SOE1 positions were either accurate or within 100 bp of current annotations given by Sanger, 12% (25 genes) are between 100 and 1000 bp from the annotated positions and ONLY 6.3% (13 genes) are beyond 1000 bp downstream of the annotated position. Two anomalies were also found: ARFGAP1 and GSTT2. ARFGAP1 was mapped to Chromosome 20 by LocusLink while in the case of GSTT2, the SOE1 position retrieved from LocusLink and the gene start position annotated by Sanger differed by 18 931 nt. in length (see Discussion). In addition, the gene orientation of GSTT2 on the genomic sequence given by LocusLink was also different from Sanger's current annotation.
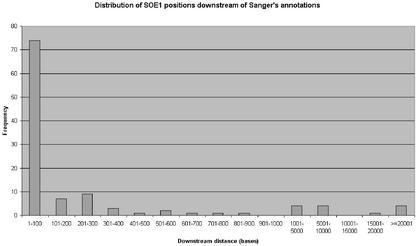
Figure 2 gives a histogram of the distribution of SOE1 positions that were given downstream of Sanger's annotated gene start positions. Table 1 shows the distribution of SOE1 positions when compared against Sanger's annotated gene start positions. The full results of the extraction for all 230 genes can be viewed on FIE2's website. A detailed table giving a comparison of SOE1 positions extracted by FIE2 for each individual gene against annotated gene start positions from Sanger is also given in the Supplementary Material for this paper. Although the positions extracted by FIE2 are given relative to the genomic contig, these contig positions were converted to chromosomal position for these 208 genes for easy reading. The calculations are based on information given at NCBI of the contig-to-chromosome positions: ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/seq_contig.md.
Figure 2.
Histogram of the distribution of SOE1 positions extracted by FIE2 (for 112 genes) which were shown to be downstream of Sanger's annotated gene start positions. The ‘Downstream distance’ indicates the difference in distance between Sanger's annotated gene start positions and FIE2's SOE1 position. This distance is measured in terms of the number of bases.
Table 1. Summary of extracted SOE1 positions from FIE2 relative to the annotated gene start positions for 168 genes of Chromosome 22 given by the Sanger Institute.
| No. of genesa | % of compared sequencesb | % of total queryc | |
|---|---|---|---|
| Extracted SOE1 position by FIE2 is the same as annotated position given by Sanger | 40 | 19.2 | 17.4 |
| Extracted SOE1 position by FIE2 is upstream of annotated position given by Sanger | 54 | 26 | 23.5 |
| FIE2's SOE1 position is downstream of annotated position given by Sanger | |||
| ≤100 bp | 74 | 35.6 | 32.2 |
| >100 bp and ≤500 bp | 20 | 9.6 | 8.7 |
| >500 bp and ≤1000 bp | 5 | 2.4 | 2.2 |
| >1000 bp | 13 | 6.3 | 5.7 |
| Anomalous annotations found between Sanger and LocusLink | 2 | 0.9 | 0.7 |
| Total | 208 | ||
aThe number of genes whose extracted 5′-most SOE1 positions were found to be either identical or upstream or downstream to the annotated Sanger gene start positions.
bThe number of genes as a percentage of all 208 genes that were compared with Sanger's annotations.
cThe number of genes as a percentage of all 230 genes that were submitted to FIE2 for extraction.
DISCUSSION
FIE2: the program and its capabilities
FIE2 has proven to be more effective than FIE version 1 (9) in extracting accurate information on the SOE1 and TIS of a gene (described below). This improved performance is due mainly to its ability to filter out irrelevant gene sequence alignments on LocusLink's EV page when more than one gene is aligned in a genomic region. An added feature of FIE2 is its ability to provide users with multiple SOE1 positions based on alignment of various mRNA transcript sequences on the chromosomal sequence. RefSeq along with other supporting sequences are used to verify the genetic locus on the contig in LocusLink. However, the authors of DBTSS (database of transcriptional start sites: http://dbtss.hgc.jp/samp_home.html), using the oligo-capping method for creating cDNA libraries, found that about a third of the RefSeqs are not 5′ end complete (10–12). The current version of LocusLink does, in fact, include the use of full-length cDNAs from the NEDO human cDNA sequencing project (denoted by the prefix ‘FLJ’) (generated by the ‘oligo-capping’ method) (13; Sugano: http://www.nedo.go.jp/bio-e/index_syokai.html) and large cDNAs (>4 kb) of the Kazusa human cDNA sequencing project (denoted by the prefix ‘KIAA’) (generated by conventional methods) (14,15). In addition, cDNA sequences from the German Cancer Research Center (DKFZ) (16,17) and IMAGE Consortium (18) are also used by LocusLink. A good number of the FLJ, KIAA, DKFZ and IMAGE cDNAs are uncharacterized and FIE2 needs to make an ‘educated guess’ as to whether these sequences represent the gene in question. If these cDNA clones bear some identity to the 5′ end of a known sequence of the gene, the sequence is considered to represent the gene in question. This cDNA sequence is then labeled as an ‘AVA’. The 5′-most aligned position of this AVA sequence on the contig is then given as a suggested SOE1 of the gene (Fig. 1A and B).
New gene annotations have recently been released for human chromosome 22 by the Wellcome Trust Sanger Institute (8). Under these new annotations, there are 393 mRNA sequences of protein coding genes which are considered complete and mapped to the genomic sequence for chromosome 22 (http://www.sanger.ac.uk/cgi-bin/hgp/chr22/display?Chr22.3.1b.coding_genes.gff). To provide a benchmark for FIE2, we performed a retrieval of SOE1 information using FIE2 and compared our results with the new annotations from Sanger. Using the names in the ‘Locus’ field of the Chr22.3.1b.coding_genes.gff file, we searched for their corresponding entries in LocusLink. However, of the 393 ‘complete’ genes annotated by Sanger, only 230 matched entries in LocusLink. The remaining 163 ‘complete’ genes annotated by Sanger were named with an accession number which was not recognized by LocusLink, for example, ‘Em:AC005500.C22.3’ (with the description ‘Matches EST sequences’). We therefore used only these 230 ‘complete’ genes to gauge FIE2's effectiveness against the current available annotations from Sanger. FIE2 was able to determine the SOE1 positions for 208 genes based on LocusLink's EV page. The SOE1 positions for the remaining 22 genes were either tagged as ‘indeterminate’ by FIE2 or could not be retrieved because there was no EV page or an incomplete EV page. Comparing the SOE1 positions of the 208 genes with the new annotations from Sanger, we found the following:
For 40 genes, the 5′-most SOE1 positions identified by FIE2 were identical to current annotations from Sanger.
The 5′-most SOE1 position for 54 genes was extended upstream of the annotated Sanger gene start position.
The 5′-most SOE1 position for 112 genes was found to be downstream of the annotated Sanger gene start position.
The information retrieved by FIE2 for two genes (ARFGAP1 and GSTT2) did not agree with Sanger's annotations.
The search results for all 230 ‘complete’ genes can be viewed on FIE's web site (http://sdmc.lit.org.sg/FIE2.0/test-dataset.html).
In the case of the two genes, ARFGAP1 and GSTT2, we found that ARFGAP1 had, in fact, been mapped to Chromosome 20 by LocusLink. As for GSTT2, the SOE1 position retrieved by FIE2 and the gene start position annotated by Sanger differed by 18 931 nt in length and the gene orientation on the genomic sequence was also in contention. We are in no position to comment on the discrepancy presented by these two genes.
For those 112 genes whose 5′-most SOE1 positions were downstream of annotated Sanger gene start positions, we found that the difference between FIE2's and Sanger's position did not exceed 100 nt in length for 74 genes. For 20 genes, the 5′-most SOE1 position were found to be downstream by between 100 and 500 nt of Sanger's annotated gene start position. Therefore, 90.4% (188 of 208 genes) of the extracted SOE1 positions were either accurate or within 500 bp of current annotations given by Sanger.
For the genes which had their 5′-most position extended upstream of the Sanger's annotated gene start position, these extensions were based on one of the following.
On RefSeqs or other representative mRNA sequences of the gene of concern: for example, the SOE1 position for MPST (LID: 4357) could be extended 4395 nt upstream of Sanger's annotated position based on the alignment of its RefSeq sequence, NM_021126 on the genomic sequence.
On ‘AVA’s which bore a high degree of identity to the gene of concern in the 5′ end of the sequence: for example, in the case of ARHGAP8 (LID: 23779), the 5′-most position given by FIE2 is based on an alignment of a NEDO sequence, AK091884 (an ‘AVA’). This AVA bears a high degree of identity to two mRNA sequences for the gene, AF195968 and AK000192 (defined as ‘FLJ20185’, an alias for ‘ARHGAP8’), over their entire length. The resulting use of the AVA, NEDO sequence AK091884, extended the 5′-most SOE1 position by 25 197 nt upstream of Sanger's start position. The description of the AVA is given alongside its accession on FIE2's information page, in this case: ‘AK091884 (H.sapiens cDNA FLJ34565 fis, clone KIDNE2006210, moderately similar to Rho GTPase activating protein 8)’. It is therefore up to the user to evaluate as to whether these AVAs are extended sequences of the gene of interest, perhaps a yet-to-be-characterized transcript variant or an entirely different gene.
However, in each case, the user can be assured that the SOE1 position is determined as a result of the alignment of an experimentally derived mRNA sequence against the genomic sequence.
The authors did, in fact, find that the 5′-most SOE1 position for BCR (LID: 613) was wrongly extended upstream of the annotated Sanger's position. The 5′-most SOE1 position was wrongly identified by FIE2 because the EV page contained an alignment using M64437. M64437's GenBank record defines it simply as ‘Human BCR mRNA, 5′ end’. The M64437 sequence is actually a sequence containing the BCR promoter (19). Therefore, this BCR sequence begins beyond and upstream of the BCR TSS. The 5′-most SOE1 position mistakenly identified by FIE2 extended the 5′ end by 155 nt upstream. However, it should be noted that the correct SOE1 position was also identified by FIE2, based on the alignment of the mRNA sequences, NM_021574 and Y00661, on the contig and this is IDENTICAL to the annotated Sanger gene start position.
The main reasons for FIE2's inability to extract information of the SOE1 or TIS position is usually due to either:
the lack of an EV page in LocusLink (that is to say, no sequence alignment on the respective genomic contig was carried out for the gene in question); or
the 5′ end of the gene sequence falls within a gapped region of the chromosome (a region where the genomic sequence has not been elucidated).
FIE2's name recognition ability has been greatly enhanced (over FIE version 1) as certain adjectives/terminologies are no longer recognized by FIE2 as being part of the gene name or symbol. However, its name recognition module can be further improved to recognize subtle nuances without comprising on its speed. For example, given the gene name LIMK2, FIE2 is currently unable to tell that the names, LIMK-2/LIMK 2, also represent the sequence. Furthermore, FIE2 is also able to class SOE1 positions deemed to be indeterminate into four separate categories (a detail explanation of these four categories is give on FIE2's FAQ page at http://sdmc.lit.org.sg/FIE2.0/faq.html). Such detailed classification was previously not provided by FIE version 1.
Comparison of FIE2 against other similar programs
Recently, a program for the extraction of eukaryotic promoter sequences from GenBank (abbreviated to PEG), was developed (4). The similarities and differences between the two programs are as follows:
Multiple SOE1 positions are presented by FIE2 based on the determination of the position of the 5′-most end of various annotated mRNA sequences which are deemed to be representative of a gene. The set of representative mRNA sequences are preselected by NCBI's LocusLink and may sometimes include full-length, uncharacterized cDNA sequences from the NEDO human cDNA sequencing project (FLJ), Kazusa DNA Research Institute (KIAA), German Cancer Research Center (DKFZ) and IMAGE Consortium, which are highly similar to the gene in question. These FLJ, KIAA, DKFZ and IMAGE sequences are usually denoted by an accession number assigned by the individual research institute. PEG searches the 5′-most mRNAs of the gene of concern by iteratively extending mRNA sequences at the 5′ end and it is possible that such cDNA sequences (FLJ, KIAA, DKFZ, IMAGE) could be omitted by PEG.
Sequences extracted by PEG can only go as far upstream as is annotated in GenBank's record, and thus cannot be directly extended further upstream. FIE2 does not have this limitation since the sequence extraction in our program is based on currently available human genome working draft sequences.
FIE2 is able to identify the TIS position of a gene and extract the sequence around it, while PEG does not have this functionality. In some cases, recognition of multiple TIS positions is possible where transcript variants for a particular gene are identified [for example, the gene ADSL (LocusID: 158) on chromosome 22 has two possible open reading frames (ORFs): GI:28904 and GI:28905].
Currently, FIE2 only supports the extraction of human sequences, but PEG can extract sequences from a broader spectrum of organisms (eukaryotes).
Both PEG and FIE2 attempt to extract the promoter region based on currently available mRNA sequences—in the case of PEG, it does so from GenBank's records, while for FIE2, it does so based on curated RefSeq and other supporting mRNA sequences which LocusLink has identified and aligned against the genomic contig.
It has to be highlighted that, for both FIE2 and PEG programs, there is a possibility that the 5′ end of the mRNA sequence may be incomplete but that does not negate the importance of the information extracted by these two programs. Both PEG and FIE2 try to make the best use of currently available information. Although the methodology and functionality of the two programs might differ, the aims of both programs are similar: to try and extract a length of sequence around what might be the promoter region based on currently available information so as to help facilitate in follow-up experiments in the lab and in silico in the studies of gene expression regulation.
The TSS is usually a good reference marker of the promoter region and it is true that only a handful of TSSs have been experimentally verified, as annotated by the Eukaryotic Promoter Database (20,21). However, both FIE2 and PEG are not trying to pinpoint the TSS, but are, instead, trying to extract a length of sequence that contains all, or part, of the promoter region (in FIE2, this depends on the length specified by the user). The promoter region can cover a region upstream of and overlapping the TSS and perhaps, extending downstream, nearing the TIS.
Theoretically, the SOE1 (‘start of exon 1’) is the TSS. However, in FIE, we use the annotation ‘SOE1’ loosely because the position, as given on LocusLink, may not sometimes be the true TSS but rather the 5′-most aligned position of an mRNA sequence on the genomic contig. For example, the mRNA sequences for the gene of concern may be 5′-incomplete or the alignment of mRNA sequences on the genomic sequence may not always provide a match with high identity in the 5′ end. Thus, the 5′-most position of the alignment on the genomic sequence may not represent the true starting point of exon 1.
A second program, EZ-Retrieve, aims to retrieve the promoter region of genes using LocusLink's Abstract-Syntax-Notation-One (ASN.1) annotation file to obtain the gene's coordinates on the contig (5). However, this gives only an approximation of the gene start position since the start coordinate given in the ASN.1 files refers to the locus and not the gene because LocusLink is, after all, locus-oriented. Two key differences between FIE2 and EZ-Retrieve can be summerized as follows:
Where FIE2 gives the users multiple SOE1 positions based on the alignment of a set mRNA sequences, in some cases, transcript variants representative of a gene, EZ-Retrieve identifies the approximate start position of a gene based on the locus coordinates and presents the user with a single approximate gene start position.
FIE2 is able to identify the TIS position for a gene whereas this feature is not available in EZ-Retrieve.
We also carried out a similar extraction with FIE version 1 to make a fair comparative analysis of the original program with the current FIE2 program. We found that FIE version 1 could only locate the SOE1 positions for 201 genes. However, on closer look, we found that the SOE1 positions for 19 out of the 201 genes differed from those extracted by FIE2. For example, the SOE1 position for ECGF1 (LocusID: 1890) wrongly identified at position 105 989 on the contig NT_011526.4 by FIE version 1 when, in fact, the 5′-most position should have been at position 105 455 based on an alignment of NM_001953 on the contig. This is due to the fact that FIE version 1 takes the 5′-most position of the contig presented on the EV page when the EV page states that there is ‘1 gene found in this genomic region’. It should be remembered that FIE version 1 will only process the EV page if there is only one gene in the genomic region presented (9). In cases where the EV page states that there is >1 gene in the presented genomic region, then FIE version 1 does not attempt to extract any information of the SOE1 or TIS position (9). However, in the case of ECGF1, in spite of the fact that the EV pages states that there is only ‘1 gene found in this genomic region’, we see that an mRNA sequence, S72487 [GenBank description: orf1 5′ to PD-ECGF/TP…orf2 5′ to PD-ECGF/TP (human, epidermoid carcinoma cell line A431, mRNA, 3 genes, 1718 nt)], was aligned to the contig. FIE2 correctly identified NM_001953 as a valid accession and therefore gave the correct 5′-most SOE1 position while also correctly identifying that S72487 was an invalid accession based on its ability to recognize the gene name or symbol in the mRNA's GenBank description. In summary, FIE version 1 was only able to identify correctly the SOE1 position for 182 genes as compared to FIE2's 208 genes (out of the possible 230 genes found on LocusLink). The extraction carried out using FIE version 1 can be viewed on its web site (http://sdmc.lit.org.sg/FIE/test-dataset.html).
There are three major human genome online resources, namely, NCBI's Human Genome information resource (http://www.ncbi.nlm.nih.gov/genome/guide/human/) (22), Ensembl's Human Genome Browser (http://www.ensembl.org/Homo_sapiens/) (23) and the Human Genome Browser at UCSC (http://genome.ucsc.edu/) (nicknamed ‘GoldenPath’) (24). The interactive tools offered by these organizations allow researchers to view the genome at a ‘macroscopic’ level—that is, at the level of an exon–intron, a gene or a chromosomal band (as opposed to a base-by-base level). FIE2 complements these browsers by giving researchers a tool for easy extraction of the base sequence of specific genomic regions around a gene's 5′ end. With Ensembl's Human Genome Browser, a user may search for a gene and get back such information as its genomic location, similarity matches (that is, related records pertaining to the gene in HUGO, SWISS-PROT, etc.), transcript structure and protein structure. Opening another window on their computer, a user could, in theory, use Ensembl's EnsMart or ContigView to retrieve a customized length of DNA sequence and thus, seemingly perform the same functions as FIE2. However, it would be prudent to take note of the fact that although the genomic location for a particular gene is supported by comparisons to protein, cDNA and EST data, the given start coordinate of the gene is sometimes either a GeneWise or Genscan prediction. FIE2 ‘reads and interprets’ the sequence alignments of representative mRNAs on the contig and then extracts and presents all information, based on its analysis, in a concise form. In effect, FIE2 provides an extension of LocusLink by streamlining the extraction of genomic sequence around a gene's 5′ end. In some cases, FIE2 refines and reorganizes the LocusLink data to supply the user with more reliable information. This one-shot analysis and processing by FIE2 thus helps the user save valuable time and effort.
Coleman and colleagues (25) estimated that the alignment of reference mRNAs to genomic sequence allows promoters to be identified for at least 75% of genes. This, therefore, lends support to the concept on which FIE2 in based on. The results for FIE2 are very promising and show definitively that the new algorithm for FIE2 is a vast improvement over that of the older version of FIE.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
REFERENCES
- 1.Maglott D.R., Katz,K.S., Sicotte,H. and Pruitt,K.D. (2000) NCBI's LocusLink and RefSeq. Nucleic Acids Res., 28, 126–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pruitt K.D., Katz,K.S., Sicotte,H. and Maglott,D.R. (2000) Introducing RefSeq and LocusLink: curated human genome resources at the NCBI. Trends Genet., 16, 44–47. [DOI] [PubMed] [Google Scholar]
- 3.Pruitt K.D. and Maglott,D.R. (2000) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang T. and Zhang,M. (2001) Promoter Extraction from GenBank (PEG): automatic extraction of eukaryotic promoter sequences in large sets of genes. Bioinformatics, 17, 1232–1233. [DOI] [PubMed] [Google Scholar]
- 5.Zhang H., Ramanathan,Y., Soteropoulos,P., Recce,M.L. and Tolias,P.P. (2002) EZ-Retrieve: a web-server for batch retrieval of coordinate-specified human DNA sequences and underscoring putative transcription factor binding sites. Nucleic Acids Res., 30, e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bajic V.B., Chong,A., Seah,S.H. and Brusic,V. (2002) Intelligent system for vertebrate promoter recognition. IEEE Intelligent Systems, 17, 64–70. [Google Scholar]
- 7.Bajic V.B., Seah,S.H., Chong,A., Zhang,G., Koh,J.L. and Brusic,V. (2002) Dragon Promoter Finder: recognition of vertebrate RNA polymerase II promoters. Bioinformatics, 18, 198–199. [DOI] [PubMed] [Google Scholar]
- 8.Collins J.E., Goward,M.E., Cole,C.G., Smink,L.J., Huckle,E.J., Knowles,S., Bye,J.M., Beare,D.M. and Dunham,I. (2003) Reevaluating human gene annotation: a second-generation analysis of chromosome 22. Genome Res., 13, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chong A., Zhang,G. and Bajic,V.B. (2002) Information and sequence extraction around the 5′-end and translation initiation site of human genes. In Silico Biol., 2, 461–465. [PubMed] [Google Scholar]
- 10.Suzuki Y., Yamashita,R., Nakai,K. and Sugano,S. (2002) DBTSS: DataBase of human Transcriptional Start Sites and full-length cDNAs. Nucleic Acids Res., 30, 328–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamashita R., Suzuki,Y., Sugano,S. and Nakai,K. (2001) DBTSS: DataBase of Human Transcriptional Start Sites and Full-Length cDNA. Genome Informatics, 12, 488–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki Y., Yoshitomo-Nakagawa,K., Maruyama,K., Suyama,A. and Sugano,S. (1997) Construction and characterization of a full length-enriched and a 5′-end-enriched cDNA library. Gene, 200, 149–156. [DOI] [PubMed] [Google Scholar]
- 13.Maruyama K. and Sugano,S. (1994) Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene, 138, 171–174. [DOI] [PubMed] [Google Scholar]
- 14.Nagase T., Nakayama,M., Nakajima,D., Kikuno,R. and Ohara,O. (2001) Prediction of the coding sequences of unidentified human genes. XX. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro.DNA Res., 8, 85–95. [DOI] [PubMed] [Google Scholar]
- 15.Ohara O., Nagase,T., Ishikawa,K., Nakajima,D., Ohira,M., Seki,N. and Nomura,N. (1997) Construction and characterization of human brain cDNA libraries suitable for analysis of cDNA clones encoding relatively large proteins. DNA Res., 4, 53–59. [DOI] [PubMed] [Google Scholar]
- 16.Wellenreuther R., Wiemann,S., Heiss,D. and Poustka,A. (2001) Generating and sequencing full-length cDNAs of novel human genes within the German cDNA consortium. In 11th Annual Workshop: Beyond the Identification of Transcribed Sequences: Functional and Expression Analysis, Washington DC. [Google Scholar]
- 17.Wiemann S., Weil,B., Wellenreuther,R., Gassenhuber,J., Glassl,S., Ansorge,W., Bocher,M., Blocker,H., Bauersachs,S., Blum,H. et al. (2001) Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs. Genome Res., 11, 422–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lennon G., Auffray,C., Polymeropoulos,M. and Soares,M.B. (1996) The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics, 33, 151–152. [DOI] [PubMed] [Google Scholar]
- 19.Shah N.P., Witte,O.N. and Denny,C.T. (1991) Characterization of the BCR promoter in Philadelphia chromosome-positive and -negative cell lines. Mol. Cell. Biol., 11, 1854–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Praz V., Perier,R., Bonnard,C. and Bucher,P. (2002) The Eukaryotic Promoter Database, EPD: new entry types and links to gene expression data. Nucleic Acids Res., 30, 322–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perier R.C., Praz,V., Junier,T., Bonnard,C. and Bucher,P. (2000) The eukaryotic promoter database (EPD). Nucleic Acids Res., 28, 302–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wheeler D.L., Church,D.M., Lash,A.E., Leipe,D.D., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Tatusova,T.A., Wagner,L. and Rapp,B.A. (2002) Database resources of the National Center for Biotechnology Information: 2002 update. Nucleic Acids Res., 30, 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hubbard T., Barker,D., Birney,E., Cameron,G., Chen,Y., Clark,L., Cox,T., Cuff,J., Curwen,V., Down,T. et al. (2002) The Ensembl genome database project. Nucleic Acids Res., 30, 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kent W.J., Sugnet,C.W., Furey,T.S., Roskin,K.M., Pringle,T.H., Zahler,A.M. and Haussler,D. (2002) The human genome browser at UCSC. Genome Res., 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coleman S.L., Buckland,P.R., Hoogendoorn,B., Guy,C., Smith,K. and O'Donovan,M.C. (2002) Experimental analysis of the annotation of promoters in the public database. Hum. Mol. Genet., 11, 1817–1821. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.