Abstract
In this paper we present several web-based tools to identify conserved patterns in sequences. In particular we present details on the functionality of PROMO version 2.0, a program for the prediction of transcription factor binding site in a single sequence or in a group of related sequences and, of MALGEN, a tool to visualize sequence correspondences among long DNA sequences. The web tools and associated documentation can be accessed at http://www.lsi.upc.es/~alggen (RESEARCH link).
INTRODUCTION
The sequencing of a large number of genomes has greatly stimulated the development of computational methods for the identification of signals or patterns in biological sequences. Conserved patterns, preserved during evolution, may be indicative of functionality and generate testable hypotheses. In our group we have developed a number of pattern-search algorithms and web-based tools that can assist in the discovery of biological function: TRANSPO (1), to search for miniature inverted repeats transposable elements in genomic sequences; MREPATT (Roset et al., manuscript submitted), to identify statistically meaningful consecutive repeated patterns in multiple genomes; MALGEN, to detect sequence motifs that are conserved among two or more very large sequences; and PROMO (2), to identify transcription factor binding sites in one or more sequences. In the ALGGEN web server one can also access clustering tools for DNA sequences using spanning trees (1) and an assembly program for sets of EST (expressed sequence tag) sequences. To facilitate the use of the programs we have developed very time-efficient algorithms so that, in most cases, the output can be provided online in a matter of seconds. The web interface of the different programs has been designed to be as user-friendly as possible.
Among the programs that can be accessed at our server, the present paper focuses on MALGEN and PROMO version 2.0. TRANSPO and MREPATT are extensively documented in recent publications. A first version of PROMO has also been published (2), but we have made significant improvements and added new features that justify its treatment in this paper.
PROMO VERSION 2.0
One of the most challenging aspects of genome biology is the understanding and modelling of the gene expression regulatory networks that operate in cells and tissues. The reliable identification of transcription factor binding sites in DNA sequences is an important step. To predict binding sites in sequences one can use the available information on known target sequences in regulatory regions of genes. Several databases contain collections of known binding sites, such as TRANSFAC (3), which contains the largest available collection of DNA binding sites in eukaryotes. Given the intrinsic variability of the protein recognition signals an appropriate representation of the binding sites are positional weight matrices (4), which store information of the relative frequency of different nucleotides in the recognition sites. In PROMO, weight matrices are constructed from known binding sites extracted from TRANSFAC and used for the identification of potential binding sites in sequences. A number of other programs exist for the prediction of transcription factor binding sites that use weight matrices (5,6) but PROMO contains a number of unique features. Among them we would like to highlight the following: (i) the possibility to select sites from any species or group of species of interest; (ii) the automatic construction of matrices that correspond to the selected taxonomic level; (iii) information in the output on other genes that may be similarly regulated; and (iv) the possibility to analyze and compare multiple sequences at the same time.
The first step when using PROMO is the selection of species or taxonomic level, both for factors and binding sites. This is aided by a Java applet that can be accessed from ‘SelectSpecies’ at the main menu. After the species selection, the matrices are constructed on the fly and can then be inspected using the ‘ViewMatrices’ option. Subsequently, the user can enter the sequence at the ‘SearchSites’ form page. The query sequence is scanned for sites with high similarity to the matrices (6). To optimize the search time we use an automata that contains all possible subsequences in the query sequence that score above the similarity threshold to any of the matrices. The output contains a graphical representation of the predicted binding sites, expectation values to assess the significance of the matches and a list of genes that are known to be regulated by the transcription factors that appear in the predictions, either individually or in all possible combinations. The information on other genes may be very useful, as the observation of functional relatedness between these genes and the gene under study may point to particularly relevant hits. The main PROMO menu also contains an option to visualize the specificity of matrices derived from different groups of organisms, ‘MatrixSpecificity’ (2) and a help page.
The construction of matrices in PROMO is an automated process. The algorithm finds, given n factor-specific binding site sequences, the subset of at least n/2 sequences which results in the longest number of consecutive completely conserved positions. By doing this we maximize specificity while keeping a representative number of sequences. For example, from 34 binding sequences available for the AP-1 transcription factor, we use 20 sequences to construct the matrix, as we have determined that 20 (higher than 34/2), but no more, can be aligned with five conserved positions. We recently tested the algorithm by comparing the results obtained using the factor-specific binding sites with those obtained with random sequences of the same length and composition as the binding sites considered. Matrices with the same number of conserved sites as expected by chance are rare and discarded by the program. In the example above the random model resulted in an expectation of three conserved positions. The matrix derived from the AP-1 binding sites, with five conserved positions, is clearly significant. The number of matrices depends on the species or taxonomic level selected by the user. For example, in the current version the program generates 503 matrices when all species are considered, 313 when only sites from animals are used and 163 when only sites from human sequences are used.
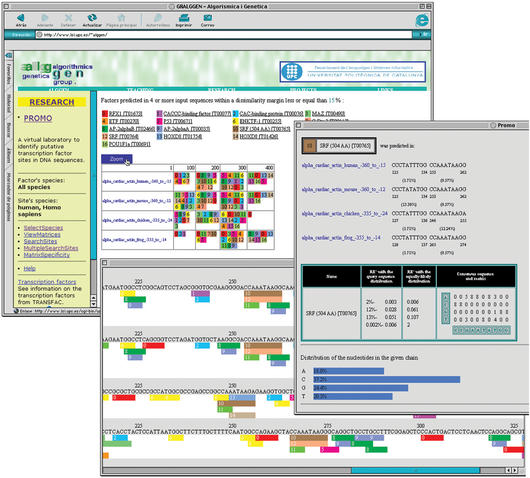
The prediction of transcription factor binding sites using weight matrices derived from collections of known sites is likely to detect the occurrence of existing sites in a sequence but will also result in the prediction of many sites which are not real, that is, false positives (5). This is a consequence of the fact that binding sites tend to be short and therefore they have a high probability of occurring by chance in any sequence. Thus, although computational prediction clearly reduces the candidate number of regulatory factors to be tested, it is not sufficient to obtain a reliable map of gene expression regulatory elements in a sequence of interest. Other biological support needs to be sought. In particular, comparative analysis of functionally related genes may provide very valuable information as these genes may share regulatory elements. Functionally related genes may be those that show similar expression patterns, as determined by array-based experiments (7) or those that have a common ancestor (orthologs). A new module of PROMO, in version 2.0, ‘MultiSearchSites’, has been designed to identify those binding sites that are present in several, or all, out of a set of user input sequences, which may for example correspond to a cluster of similarly expressed genes. For the analysis of a group of related sequences the ‘MultiSearchSites’ option, instead of the ‘SearchSites’ option, should be selected from the main menu. Parameters that can be modified by the user are the percentage of sequences that are required to contain a match to the binding site so that the match is reported and the similarity threshold used in the predictions. The requirement that several sequences must contain the match reduces the number of total predictions while keeping those that may be more relevant. An example is shown in Figure 1, where sites above 85% similarity and present in all sequences are reported. The example corresponds to the regulatory region of the cardiac alpha-actin gene from four different vertebrate species. The prediction of the SRF (serum response factor) binding site corresponds to the experimentally verified site (8).
Figure 1.
PROMO ‘MultiSearchSites’ output example. The example corresponds to the regulatory region of the cardiac alpha-actin gene from four different vertebrate species: humans, mouse, chicken and frog. Only those binding site predictions that appear in all four sequences are shown, as boxes of different colour and number. The image below, where the sequences are shown, is the result of selecting ‘Zoom’ in the main results page above. The image on the right is a detail of the SRF (serum response factor)—binding site predictions on the sequences. It also shows the weight matrix for the SRF recognition site and random expectation (RE) values for different levels of sequence-matrix similarity. The RE is calculated with a model that considers that all nucleotides are equally probable and also with a model that considers the nucleotide composition in the query sequence (in the picture represented by blue bars below matrix).
MALGEN
MALGEN is the acronym of Multiple ALignment of GENomes and it is a web tool to explore sequence relationships among large DNA sequences. Sequence segments of a minimum user-defined length present in two different sequences are identified and represented graphically (see examples in Fig. 2). Regions of identity, or matching segments, are marked with vertical lines, where green lines represent exact direct identities and red lines exact inverse identities. More than two sequences can also be represented. The identity segments are of maximum length, as they cannot be extended further, and they are unique, as they represent one-to-one correspondences, that is, matches that occur only once in each DNA sequence. For these reasons they are called Maximal Unique Matchings, for short MUMs. Note that the uniqueness property is a strong requirement but it may reinforce the biological interpretation of the matches.
Figure 2.
Comparison of three C.pneumoniae strain genomes: AR39 (1.247 Mb), CWL029 (1.248 Mb), J138 (1.175 Mb), in this order top to bottom. Horizontal white lines are the DNA sequences, vertical green lines are exact direct identities and vertical red line exact inverse identities.
The comparison of long DNA sequences is computationally expensive and not many tools for this purpose have so far been developed. Of the existing ones, MUMER v2 (9) can only compare two genomes and MGA (10) aligns multiple sequences but with a very high cost of space. We have designed an efficient space–time algorithm that allows the comparison between many genomes simultaneously. The algorithm only needs a linear space with respect to the shortest sequence. Its theoretical basis is described in detail elsewhere (11).
The web interface of MALGEN can be accessed from our web site under the Research and Align Tools links. The email of the user is required as his identifier. The user may access previously submitted jobs or, otherwise, start a new job. The process has two parts. In the first one the sequence files are provided by the user through the entry form and the server searches for the collection of MUMs, stores the list of MUMs as a new job and sends an email to the user. The list of MUMs is stored because their search is the most time-expensive process. In the second part, the user can access the existing job, containing the list of MUMs and generate a graphical representation on the fly. Different options for visualization are provided by a user-friendly interface. Options include the minimum length of the MUMs that will appear in the picture, the possibility to show direct or inverse matches, or both at the same time, the setting of the distance between consecutive sequences and the selection of the order of appearance of the sequences. The current MALGEN web tool runs the pair-wise version of the algorithm; the implementation for multiple genomes is in progress.
Depending on the nature of the sequences and the MUM minimum size, MALGEN may provide information on different kinds of signals. As the program can deal with very long sequences, it is particularly suited for the identification and visualization of chromosome, or genome, rearrangements among related species. At the same time it can provide an accurate and exhaustive mapping, in the form of MUMs, between chromosomes or genomes. Figure 2 shows a comparison between the genomes of three Chlamidophila pneumoniae strains (minimum MUMs size of 18 bases). In this case MUMs occupy 98% of the sequence. It can be observed that the superior one, which corresponds to the AR39 strain, contains two large inverse translocated segments in respect to the other two strains. Other examples can be visualized at the MALGEN site by submitting with the default email (malgen@lsi.upc.es). MALGEN can be used to identify different types of functional elements on genomes. For example, we are currently exploring its use in the identification of exons by comparative genomics. An additional use of MALGEN is in the alignment of very long sequences by using the MUMs to anchor the alignment (12).
Acknowledgments
ACKNOWLEDGEMENTS
ALGGEN group is supported by IST programme of the EU under contract IST-1999-14186 (ALCOM-FT).
REFERENCES
- 1.Santiago N., Herraiz,C., Goñi,J.R., Messeguer,X. and Casacuberta,J.M. (2002) Genome-wide analysis of the emigrant family of mites of Arabidopsis thaliana. Mol. Biol. Evol., 19, 2285–2293. [DOI] [PubMed] [Google Scholar]
- 2.Messeguer X., Escudero,R., Farré,D., Núñez,O., Martínez,J. and Alba,M.M. (2002) PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics, 18, 333–334. [DOI] [PubMed] [Google Scholar]
- 3.Matys V., Fricke,E., Geffers,R., Gossling,E., Haubrock,M., Hehl,R., Hornischer,K., Karas,D., Kel,A.E., Kel-Margoulis,O.V. et al. (2003) TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res., 31, 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bucher P. (1990) Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol., 212, 563–578. [DOI] [PubMed] [Google Scholar]
- 5.Roulet E., Fisch,I., Junier,T., Bucher,P. and Mermod,N. (1998) Evaluation of computer tools for the prediction of transcription factor binding site on genomic DNA. In Silico Biol., 1, 21–28. [PubMed] [Google Scholar]
- 6.Quandt K., Frech,K., Karas,H., Wingender,E. and Werner,T. (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acid Res., 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang M.Q. (1999) Large-scale gene expression data analysis: a new challenge to computational biologists. Genome Res., 9, 681–688. [PubMed] [Google Scholar]
- 8.Sartorelli V., Webster,K.A. and Kedes,L. (1990) Muscle-specific expression of the cardiac alpha-actin gene requires MyoD1, CArG-box binding factor, and Sp1. Genes and Dev., 4, 1811–1822. [DOI] [PubMed] [Google Scholar]
- 9.Delcher A.L., Phillippy,A., Carlton,J. and Salsberg,L. (2002) Fast algorithm for large-scale genome alignment and comparison. Nucleic Acid Res., 11, 2478–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Höhl M., Kurtz,S. and Ohlebush,E. (2002) Efficient multiple genome alignment. Bioinformatics, 18, 1–9. [DOI] [PubMed] [Google Scholar]
- 11.Huerta M. and Messeguer,X. (2002) Efficient space and time multicomparison of genomes. Research Report LSI-02-64-R, Dep. Llenguatge i Sistemes Informàtics, Universitat Politècnica de Catalunya. [Google Scholar]
- 12.Delcher A.L., Kasif,S., Fleischmann,R.D., Peterson,J., White,O. and Salsberg,L. (1999) Alignment of whole genomes. Nucleic Acid Res., 27, 2369–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]