Abstract
Phylogenetic footprinting is a method for the discovery of regulatory elements in a set of homologous regulatory regions, usually collected from multiple species. It does so by identifying the best conserved motifs in those homologous regions. This note describes web software that has been designed specifically for this purpose, making use of the phylogenetic relationships among the homologous sequences in order to make more accurate predictions. The software is called FootPrinter and is available at http://bio.cs.washington.edu/software.html.
DESCRIPTION
One of the current challenges facing biologists is the discovery of novel functional elements in non-coding genomic sequence. With the rapidly increasing number of genomes being sequenced, a comparative genomics approach called ‘phylogenetic footprinting’ (1) has become a favored method for such discovery.
This note focuses on the discovery of novel regulatory elements. The idea underlying phylogenetic footprinting is that selective pressure causes regulatory elements to evolve at a slower rate than the non-functional surrounding sequence. Therefore the best conserved motifs in a collection of homologous regulatory regions are excellent candidates as regulatory elements.
The traditional method that has been used for phylogenetic footprinting is to construct a global multiple alignment of the homologous regulatory sequences and then identify well conserved aligned regions (2). However, this approach fails if the regulatory regions considered are too diverged to be accurately aligned.
In earlier work (3,4), we described an algorithm designed specifically for phylogenetic footprinting. Instead of relying on multiple alignment, we attack the problem with a motif discovery approach. Given a set of homologous input sequences and the phylogenetic tree T relating them, the algorithm identifies every set of kmers, one from each input sequence, that have parsimony score at most d with respect to T, where k and d are parameters specified by the user. (The parsimony score is the minimum number of nucleotide substitutions along the branches of T that explain the set of identified kmers.) This algorithm has been implemented in a program called FootPrinter, available at http://bio.cs.washington.edu/software.html, both in source code and through a web interface. This note describes the web interface to FootPrinter. The reader is referred to earlier work (3–5) for details on FootPrinter's algorithm, its applications on biological data and comparison to other phylogenetic footprinting tools.
BASIC USER INPUTS
The simple web form asks the user to supply the homologous input sequences in Fasta format. The first word of each sequence annotation line following ‘>’ must correspond to the name of a species in the phylogenetic tree. The user also supplies the phylogenetic tree relating the sequences, although if the tree is absent FootPrinter will use a default species tree containing many of the most commonly used eukaryotic species. If the user chooses to enter a phylogeny, it is given in the usual bracket form. For example, the tree for Figure 1 is ((salmon,(lates,fugu)),(chicken,(((rat,mouse),human),(dog, sheep)))).
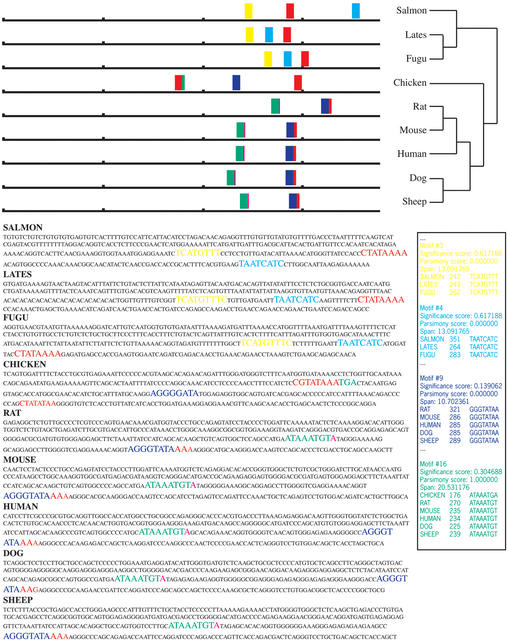
Figure 1.
Sample FootPrinter output on the upstream regions of nine growth hormone genes. We searched for motifs of size 8 with at most two mutations. Motif losses were allowed, at a cost of one mutation. The subregion size was set to 100 bp with a subregion change cost of one mutation. The motif list reported on the right has been trimmed to fit on the page.
The user also chooses a few parameter values that specify the type of motif FootPrinter should report. These include the motif size (k in the description above) and the maximum number of mutations (maximum parsimony score d in the description above).
BASIC FOOTPRINTER OUTPUT
FootPrinter's results are made available in three formats, HTML, Postscript and plain text. The HTML format is assumed in what follows, as it provides the most information in a graphical, interactive form. See Figure 1 for an example of the results obtained on a set of homologous growth hormone regulatory regions.
Each of the input sequences is repeated on the results page, with FootPrinter's discovered motifs highlighted in color and in larger fonts. Instances of the same motif appear in the same color. The font size indicates the parsimony score, with larger fonts corresponding to solutions with smaller parsimony score.
At the top of the results page the user can see a schematic representation of all the motifs at a glance. Each sequence is represented by a horizontal line labeled at one end by the species name. The phylogenetic tree relating the sequences is also shown. Above each horizontal line colored bars indicate the positions of FootPrinter's discovered motifs. The bar colors correspond to the font colors used in the sequences themselves. A more classical text representation is also available in the lower right panel.
In the example of Figure 1, the motif size was set to 8 and the maximum parsimony score set to 2. The reader will notice, however, that reported motifs are sometimes longer than the prescribed length (for example, the yellow motif in fishes is of length 9). This is because FootPrinter merges together overlapping motifs that it discovers, provided every instance overlaps in exactly the same way. Conversely, if motifs overlap differently in different sequences, each will be assigned its own color. Nucleotides that belong to several motifs are colored according to the motif with the most significant degree of conservation (see, for example, the overlapping green and purple motifs).
ADDITIONAL USER INPUTS
There are two further sets of input parameters that deserve mention. The first has to do with conservation of motif position. If the user does not want FootPrinter to report motifs whose locations within the input sequences vary too much, parameters called ‘subregion size’ (with units in bp) and ‘subregion change cost’ can be used. In this case, FootPrinter subdivides the input sequences into subregions of the given size and the given cost is charged every time a motif changes subregion during its evolution. This cost is added to the motif's parsimony score, so that, in order to be reported by FootPrinter, such motifs must be better conserved if they are not to exceed the maximum parsimony score specified by the user. A ‘soft boundary’ approach ensures that nearby motifs separated by a subregion boundary are not penalized (4).
The final set of input parameters is very important in practice. In sufficiently diverged sequences it may be common that some regulatory elements occur in only a subset of the input sequences. This could happen because the regulatory element is only functional in a subset of the sequences or also because some of the input sequences chosen happen to be too short to contain the regulatory element. In either case, it is useful for FootPrinter to allow for the loss of regulatory elements in some of the input sequences. To do so, FootPrinter starts by estimating the length of each branch of the tree based on the input sequences. The motifs reported are those whose parsimony score is unexpectedly low considering the amount of divergence of the subset of species containing the motif. If the user chooses this option, another parameter called the ‘motif loss cost’ is added to the parsimony score for every branch on which the motif is lost.
ADDITIONAL FOOTPRINTER OUTPUT
Referring again to Figure 1, we have already discussed two motifs (green and yellow) that occur in only a subset of species. Even though these motifs are absent from many species, their level of conservation and span of the tree are judged significant by FootPrinter according to criteria previously described (3). The light blue motif, found only in fishes, may be a false positive, as it is clear from the schematic representation that its position is inconsistent with respect to the other two motifs in fishes. FootPrinter leaves this judgment call to the user.
There are a few more functionalities of the HTML output format that are useful to the user. If the mouse cursor is moved to a colored instance in one of the sequences (without clicking the mouse button), information about that motif is shown at the bottom of the browser screen: the position of this instance, the parsimony score of this motif, and the total branch length spanned by the input sequences containing instances of this motif. If the mouse button is now clicked the corresponding colored bars in the schematic at the top of the page are highlighted and the subtree containing instances of the motif is colored in the phylogeny at the top of the page. This allows for quick visual identification of motif occurrences. A textual summary of the motif clicked is also reported (lower right panel in Fig. 1).
ONLINE HELP
At every step of the process there are links to relevant information to help the user. These include definitions of input parameters, examples of input format, guidance on parameter choices and advice on parameters to change if the user would like more or fewer motifs to be reported.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Saurabh Sinha for his help in developing the web interface and an anonymous referee who tested the interface carefully. This material is based upon work supported in part by a Natural Sciences and Engineering Research Council of Canada (NSERC) fellowship, by a Fonds Québécois de la Recherche sur la Nature et les Technologies fellowship, by the Howard Hughes Medical Institute, by the National Science Foundation under grants DBI-9974498 and DBI-0218798, and by the National Institutes of Health under grant HG02602-01.
REFERENCES
- 1.Tagle D., Koop,B., Goodman,M., Slightom,J., Hess,D. and Jones,R. (1988) Embryonic ɛ and γ globin genes of a prosimian primate (Galago crassicaudatus); nucleotide and amino acid sequences, developmental regulation and phylogenetic footprints. J. Mol. Biol., 203, 439–455. [DOI] [PubMed] [Google Scholar]
- 2.Duret L. and Bucher,P. (1997) Searching for regulatory elements in human noncoding sequences. Curr. Op. Struct. Biol., 7, 399–405. [DOI] [PubMed] [Google Scholar]
- 3.Blanchette M., Schwikowski,B. and Tompa,M. (2002) Algorithms for phylogenetic footprinting. J. Comput. Biol., 9, 211–223. [DOI] [PubMed] [Google Scholar]
- 4.Blanchette M. and Tompa,M. (2002) Discovery of regulatory elements by a computational method for phylogenetic footprinting. Genome Res., 12, 739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanchette M., Kwong,S. and Tompa,M. (2003) An empirical comparison of tools for phylogenetic footprinting. In Third IEEE Symposium on Bioinformatics and Bioengineering, IEEE Press, Los Alamitos, CA, pp. 69–78. [Google Scholar]