Abstract
SEARCHPKS is a software for detection and analysis of polyketide synthase (PKS) domains in a polypeptide sequence. Modular polyketide synthases are unusually large multi-enzymatic multi-domain megasynthases, which are involved in the biosynthesis of pharmaceutically important natural products using an assembly-line mechanism. This program facilitates easy identification of various PKS domains and modules from a given polypeptide sequence. In addition, it also predicts the specificity of the potential acyltransferase domains for various starter and extender precursor units. SEARCHPKS is a user-friendly tool for correlating polyketide chemical structures with the organization of domains and modules in the corresponding modular polyketide synthases. This program also allows the user to extensively analyze and assess the sequence homology of various polyketide synthase domains, thus providing guidelines for carrying out domain and module swapping experiments. SEARCHPKS can also aid in identification of polyketide products made by PKS clusters found in newly sequenced genomes. The computational approach used in SEARCHPKS is based on a comprehensive analysis of various characterized clusters of modular polyketide synthases compiled in PKSDB, a database of modular polyketide synthases. SEARCHPKS can be accessed at http://www.nii.res.in/searchpks.html.
INTRODUCTION
Polyketides are a group of secondary metabolites exhibiting remarkable diversity both in terms of their structure and function. These metabolites have been characterized in a wide range of organisms. Many of the polyketides are clinically valuable anti-microbial, anti-fungal, anti-parasitic, anti-tumor and immunosuppressive agents. The biosynthesis of polyketides is catalyzed by a collection of enzyme activities called polyketide synthases (PKSs). These proteins utilize thioesters of acetate and other short carboxylic acids to perform sequential decarboxylative condensations followed by associated reductive reactions to produce diverse polyketide products. The catalytic domains which harbor the active sites present in PKSs have been defined on the basis of their function. An acyltransferase domain (AT) for extender unit selection and transfer, an acyl carrier protein (ACP) with a phosphopantetheine swinging arm for extender unit loading and a ketoacyl synthase (KS) domain for decarboxylative condensations are the core domains. Additional domains have been identified for the modification of the initial carbonyl group, such as, a ketoreductase (KR), a methyl transferase (O-MT), a dehydratase (DH), an enoyl reductase (ER) and an acyl CoA ligase (AL). During the biosynthesis, the growing polyketide chain remains covalently attached to the enzyme and a thioesterase (TE) domain catalyzes the release of the polyketide product, when it reaches its full length (1,2 and other reviews in the thematic issue of Chemical Reviews 1997, Vol. 97). The segments of polypeptide chain connecting all these domains are referred to as linkers and they have been shown to establish functional communication between and within modules (3).
Modular polyketide synthases catalyze the biosynthesis of polyketides through an assembly-line mechanism. These enzymes harbor sets of distinct active sites termed modules for catalyzing each condensation and chain elongation step (4). The number of modules present in a modular PKS cluster directly correlates with the number of ketide units present in the polyketide product. The domains present in each module dictate the chemical moiety which the given module would add to a growing polyketide chain. This modular logic of biosynthesis has been exploited to produce several novel compounds which have highlighted the functional versatility of these multienzyme assemblies (5,6). Most of these studies have been carried out using empirical gene fusion approaches and have invariably resulted in low product titer during fermentation. Moreover, the reported genetic manipulation experiments have been carried out with a few PKS gene clusters. With rapid increase in the number of PKS gene clusters in sequence databases, it is essential to carry out a systematic analysis of PKSs to harness their vast potential of combinatorial biosynthesis. Identification of PKS domains along with their substrate specificity and catalytic activity from a polypeptide sequence would assist in rational design of novel polyketides. Such an automated computational tool would also assist in deciphering polyketide products biosynthesized by PKS gene clusters located in the newly sequenced genomes. Although the sequences of various multifunctional PKS proteins are available in databases like SWISS-PROT or GenBank (7,8), the organization of various PKS modules or functional domains in such proteins have not been comprehensively annotated. The standard domain identification tools like Conserved Domain Database (CDD) search (9) fail to detect some of the key functional domains in PKS proteins. This inspired us to develop SEARCHPKS, a software for detection and analysis of PKS domains. This program facilitates easy identification of various PKS domains and modules from a given polypeptide sequence. In addition it predicts the specificity of the potential acyltransferase domains for various starter and extender precursor units. SEARCHPKS is a user-friendly tool for correlating polyketide chemical structures with the organization of domains and modules in the corresponding modular PKSs. This program also allows the user to extensively analyze and assess the sequence homology of various polyketide synthase domains, thus providing guidelines for carrying out domain and module swapping experiments. The computational approach (10) used in SEARCHPKS is based on a comprehensive analysis of various characterized clusters of modular polyketide synthases compiled in PKSDB, a database of modular polyketide synthases.
FEATURES OF SEARCHPKS
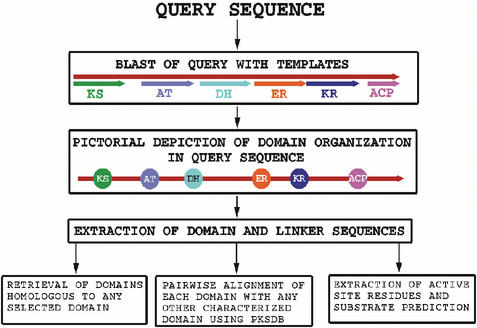
Figure 1 shows a flowchart depicting various features of SEARCHPKS. This software can identify various PKS domains from a given query sequence by using an automated computational protocol (10). The results are displayed as a pictorial representation of the domain organization. The program also provides appropriate interfaces to carry out a number of different analyses for each of the depicted domains.
Figure 1.
Flowchart depicting various features of SEARCHPKS.
Domain identification and pictorial depiction of domain organization
A major component of SEARCHPKS is the automated computational protocol for correct identification of various PKS domains in a polypeptide sequence (10). Domain identification is carried out by pairwise sequence alignment of the query sequence with template sequences of KS, AT, DH, ER, KR, TE and ACP domains. Sequences are aligned using a local version of the BLAST program (11) downloaded from the NCBI site. BLOSUM62 scoring matrix and default values for gap penalties are used for sequence alignments and only alignments having E-value <0.000001 are considered as statistically significant hits. Template sequences of KS, AT, DH, ER and KR domains have been taken from module 4 of erythromycin, while the template for TE is from module 6 of erythromycin. The boundaries of these template sequences are chosen based on the sequence analysis by Donadio and Katz (12). It may be noted that various PKS domains identified as per the boundaries suggested by Donadio and Katz (12) have been used in a number of domain swapping experiments. Thus it was considered appropriate to choose template domains based on their work. However, detailed sequence analysis (10) of various experimentally characterized PKS clusters indicated that, using the ACP domain of erythromycin module 4 as template and E-value cut-off of 0.000001, several functional ACP domains could not be detected. Therefore, for the identification of the ACP domains, a representative set of 73 diverse sequences of ACP family taken from Pfam database (13) is used as templates in SEARCHPKS program. It may be noted that the standard method for identification of various functional domains in a protein is to use CDD search. Our earlier analysis (10) of domain organization in 19 characterized modular PKS clusters has indicated that CDD search fails to detect any DH domains and cannot distinguish between KR and ER domains. SEARCHPKS correctly detects all the reductive domains by using appropriate DH, ER and KR templates. Correct identification of reductive domains is essential for predicting the chemical structure of the final polyketide product. Therefore, SEARCHKS is more useful than CDD search for the purpose of identification of polyketide reductive domains and correlating them with their polyketide product.
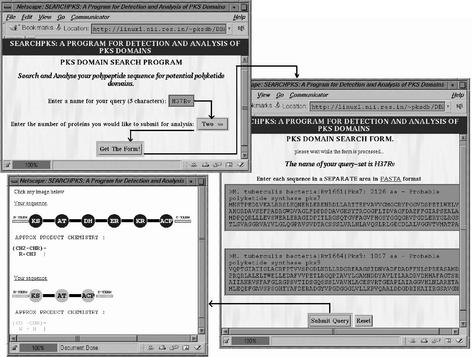
After the identification of the boundaries of various domains in the query sequence, SEARCHPKS depicts the arrangement of domains and linkers in a pictorial format with clickable links leading either to their amino acid sequences in FASTA format or for further analysis involving that domain. The modular organization is highlighted by using different colors for different modules in a potential PKS cluster. Since, PKS clusters often consist of multiple ORFs, SEARCHPKS provides options for submitting up to 10 different polypeptide sequences in a single query, so that the domain organization of an entire cluster can be viewed together in a single output. Figure 2 shows a typical result from SEARCHPKS for the ORFs Rv1661 and Rv1664 from the genome of Mycobacterium tuberculosis strain H37Rv. As can be seen, the ORF Rv1661 contains a complete module consisting of all the reductive domains, while ORF Rv1664 is a minimal module containing only the KS, AT and ACP domains.
Figure 2.
A typical use of SEARCHPKS for predicting domain organization in two ORFs Rv1661 and Rv1664 from M.tuberculosis H37Rv. On entering a five letter name for the query and selecting TWO as the number of ORFs, the program displays a form for submitting the sequences of the two ORFs in FASTA format. Upon submitting the query, the program gives a pictorial depiction of the domain organization in these two ORFs. The domains are depicted as filled circles with names of the domains inscribed in them, while the linker regions are represented as filled or shaded lines.
Extraction of sequences of various domains/linkers and their homology assessment
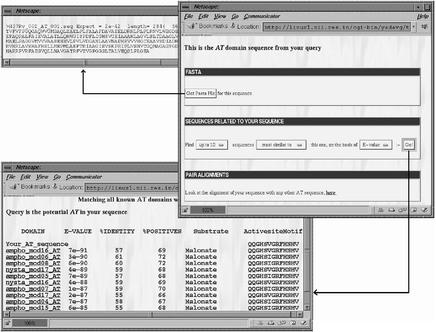
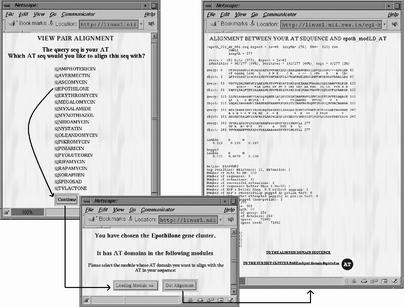
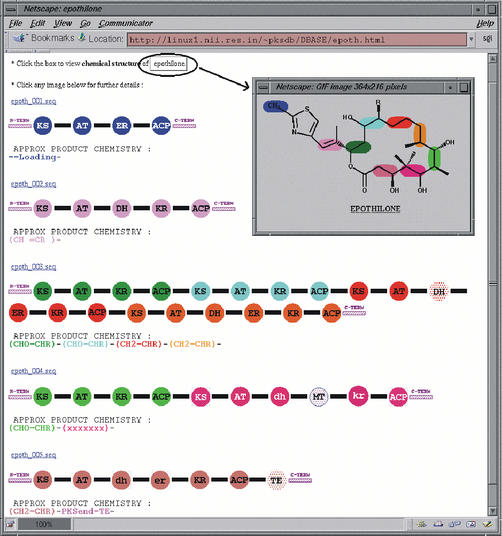
SEARCHPKS provides a very convenient interface for extracting sequences of various domains and linkers in FASTA format. Each of the PKS domains identified in the query sequence can also be compared with other similar domains found in 19 characterized PKS clusters, analyzed in detail in our earlier work (10). These 19 PKS clusters have a total of 182 KS, 188 AT, 108 DH, 29 ER, 165 KR, 192 ACP and 17 TE domains. The sequences of all these domains have been stored in a database (PKSDB) which is accessible from the query interface of SEARCHPKS. Upon selecting the option to obtain sequences of homologous domains, the query domain is aligned with every other characterized domain in PKSDB. All the pairwise alignments are then stored by SEARCHPKS in a temporary directory and the user can retrieve a specified number of sequences which are most similar or most diverse from the query domain. For the ORF Rv1661 from M.tuberculosis, Figure 3 shows a typical example of the usage of SEARCHPKS for extraction of the sequence of an AT domain in FASTA format and retrieval of 10 AT domains most similar to this AT domain. The homologous domains given in this output are in fact clickable links leading to the alignment of the query domain with each of these domains. The program also provides an option for aligning the query domain with any specific domain from the 19 characterized PKS clusters stored in PKSDB. Figure 4 shows an example where the query AT domain has been aligned with an AT domain in the loading module of epothilone PKS. From the alignment page, the user can go to the page depicting the complete domain organization of the PKS cluster from which the subject domain has been selected as well as the chemical structure of the corresponding polyketide product. For example, on clicking the link to the subject cluster in Figure 4, the user can access the page containing the domain organization of epothilone cluster along with the chemical structure of epothilone (Fig. 5). This page, accessed from PKSDB, has also been generated using SEARCHPKS and has been appropriately annotated to compare the predicted domain organization with the experimentally validated domain organization of epothilone cluster (14). This option of SEARCHPKS is useful for finding out the type of polyketide products made by homologous domains in various experimentally characterized PKS clusters.
Figure 3.
Screen dumps showing usage of SEARCHPKS for extracting the sequence of an AT domain in FASTA format and retrieving 10 AT domains most similar to the AT domain in query sequence. On clicking the AT domain of the second ORF in the screen shown in Figure 2, the program displays a page with a button leading to sequence of this domain in FASTA format and a form for extracting from PKSDB, a specified number of AT domains most similar or most diverse from the query domain. For these homologous AT domains, the program lists the degree of similarity to the query, their active site motif as well as substrate specificity along with links to their alignment with the query.
Figure 4.
Screen dumps showing alignment of the query AT domain with the AT domain in the loading module of epothilone PKS. On selecting the pair alignment option in the screen shown in Figure 3, the program displays a list of PKS clusters and upon selection of a PKS cluster, the program prompts the user to select the module from which the specified domain is to be chosen for alignment. After getting the required user input, the program displays the alignment page and this page also provides links to the modular PKS cluster from which the subject domain has been selected.
Figure 5.
Pictorial depiction of domain organization for epothilone biosynthetic cluster. All domains in the same module have been depicted using a single color, while different modules have different colors. In the chemical structure, each chemical moiety has been depicted using the same color as the color of the corresponding module of the epothilone cluster which adds it to the growing polyketide chain during biosynthesis of epothilone. Domains which have been correctly predicted by SEARCHPKS have been represented as solid filled circles, while domains which are not predicted by SEARCHPKS have been depicted as dotted filled circles. Catalytically active domains have been depicted using upper case alphabets, while names of the domains have been inscribed in lower case for catalytically inactive domains. The methyl transferase domains have been manually annotated as the software does not include templates for detection of this domain.
Substrate specificity of AT domains
Since AT domains are known to control the specificity for various starter and extender units during polyketide biosynthesis, SEARCHPKS also extracts the key active site residues of AT domains and based on the pattern of these active site residues it attempts to predict their substrate specificity. For each query AT domain, 13 active site residues are extracted from its alignment with the crystal structure of acyltransferase from Escherichia coli FAS (1MLA) (15). The choice of these 13 active site residues is based on our detailed sequence analysis and molecular modelling calculations, which indicated that the substrate specificity of AT domain is controlled by only few residues in the active site cavity (10). If the 13 active site residues of a query AT domain show an identical match to the corresponding residues in any AT domain of known specificity in our data set from 19 characterized modular PKS clusters, the query domain is assigned the same specificity as that of the matched AT domain. SEARCHPKS also lists the 13 active site residues as well as the known specificities of AT domains homologous to the query. The 13 active site residues of the AT domain in Rv1664 show exact match with several malonate specific AT domains (Fig. 3) and, as can be seen from Figure 2, this AT domain is predicted to be specific for malonate. Similarly, the AT domain in Rv1661 is predicted to be specific for methylmalonate (Fig. 2). In case no exact match of 13 active site residues is found for the query domain, SEARCHPKS only lists the 13 active site residues and the user can draw inferences about the substrate specificity by comparing this motif with the active site residues of the homologous domains.
Information about the chemical moiety added by a given PKS module
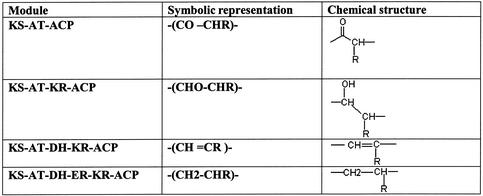
For modular PKSs, based on the type of reductive domains and specificity of the AT domain, SEARCHPKS attempts to predict the approximate chemical formulae of the moiety likely to be incorporated by a given module in the query sequence. Since SEARCHPKS uses an automated computational approach for prediction of chemical structure, for computational convenience, the chemical formulae are represented in a seven character symbolic form as shown in Table 1. With appropriate tools, these symbolic representations can be converted to conventional chemical structures.
Table 1. Symbolic representation of chemical structures.
R can be H or CH3 or any other chemical group depending on whether the AT domain is specific for malonate, methylmalonate or any other substrates.
SCOPE AND FUTURE DEVELOPMENT
SEARCHPKS is a powerful tool for the correct identification of PKS domains in a polypeptide sequence and their detailed analysis. This software uses a knowledge-based approach for prediction of domain organization and substrate specificity, based on a detailed analysis of a large collection of well annotated sequences of domains and linkers from 19 characterized modular PKS clusters (10). SEARCHPKS permits comparison of PKS domains in terms of their sequence similarity, substrate specificity and active site motifs. These features of SEARCHPKS make this software a valuable resource which can aid in identification of polyketide products biosynthesized by PKS clusters found in newly sequenced microbial genomes. Apart from analyzing the domain organization in uncharacterized PKS clusters, SEARCHPKS also permits analysis of PKS domains present in various characterized modular PKS clusters in terms of their sequence similarity and substrate specificity. Thus this software can also provide useful guidelines for designing genetic manipulation experiments to engineer novel natural products.
The present version of SEARCHPKS analyzes active site residues only for the AT domain. In the future versions of the program appropriate interfaces will be added for analysis of active site residues for other domains. This will help in identification of non-functional reductive domains. SEARCHPKS has been developed based on extensive analysis of modular PKS clusters. Even though it can predict the domain organization and the specificity of the AT domain in an iterative PKS clusters, the present version of the program cannot distinguish between modular and iterative PKS clusters. Addition of appropriate predictive rules based on further sequence/structural analysis of various characterized PKS clusters, will enhance the ability of SEARCHPKS to correlate sequence of PKS proteins to the chemical structure of their polyketide product.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr Sandip K. Basu for his encouragement and support. G.Y. is a Senior Research Fellow of CSIR, India. R.S.G. is a Wellcome Trust International Senior Research Fellow for Biomedical Science in India. This work was supported by grants to the National Institute of Immunology from the Department of Biotechnology, Government of India. Computational resources provided under BTIS project of DBT, India are gratefully acknowledged.
REFERENCES
- 1.Gokhale R.S. and Tuteja,D. (2001) Biochemistry of polyketide synthases. Biotechnology, 10, 341–372. [Google Scholar]
- 2.Hopwood D.A. (1997) Genetic contributions to understanding polyketide synthases. Chem. Rev., 97, 2465–2498. [DOI] [PubMed] [Google Scholar]
- 3.Gokhale R.S. and Khosla,C. (2000) Role of linkers in communication between protein modules. Curr. Opin. Chem. Biol., 4, 22–27. [DOI] [PubMed] [Google Scholar]
- 4.Staunton J. and Weissman,K.J. (2001) Polyketide biosynthesis: a millennium review. Nature Prod. Rep., 18, 380–416. [DOI] [PubMed] [Google Scholar]
- 5.Khosla C., Gokhale,R.S., Jacobsen,J.R. and Cane,D.E. (1999) Tolerance and specificity of polyketide synthases. Annu. Rev. Biochem., 68, 219–253. [DOI] [PubMed] [Google Scholar]
- 6.Katz L. and McDaniel,R. (1999) Novel macrolides through genetic engineering. Med. Res. Rev., 19, 543–558. [DOI] [PubMed] [Google Scholar]
- 7.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J., Rapp,B.A. and Wheeler,D.L. (2002) GenBank. Nucleic Acids Res., 30, 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchler-Bauer A., Panchenko,A.R., Shoemaker,B.A., Thiessen,P.A., Geer,L.Y. and Bryant,S.H. (2002) CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res., 30, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yadav G., Gokhale,R.S. and Mohanty,D. (2003) Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J. Mol. Biol., 328, 335–363. [DOI] [PubMed] [Google Scholar]
- 11.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 12.Donadio S. and Katz,L. (1992) Organization of the enzymatic domains in the multifunctional polyketide synthase involved in erythromycin formation in Saccharopolyspora erythraea. Gene, 111, 51–60. [DOI] [PubMed] [Google Scholar]
- 13.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam Protein Families Database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molnar I., Schupp,T., Ono,M., Zirkle,R.E., Milnamow,M., Nowak-Thompson,B., Engel,N., Toupet,C., Stratmann,A., Cyr,D.D. et al. (2000) The biosynthetic gene cluster for the microtubule-stabilizing agents epothilones A and B from Sorangium cellulosum So ce90. Chem. Biol., 7, 97–109. [DOI] [PubMed] [Google Scholar]
- 15.Serre L., Verbree,E.C., Dauter,Z., Stuitje,A.R. and Derewenda,Z.S. (1995) The E.coli malonyl CoA: acyl carrier protein transacylase at 1.5 Å resolution. Crystal structure of a FAS component. J. Biol. Chem., 270, 12961–12964. [DOI] [PubMed] [Google Scholar]