Abstract
The simple fact that proteins are built from 20 amino acids while DNA only contains four different bases, means that the ‘signal-to-noise ratio’ in protein sequence alignments is much better than in alignments of DNA. Besides this information-theoretical advantage, protein alignments also benefit from the information that is implicit in empirical substitution matrices such as BLOSUM-62. Taken together with the generally higher rate of synonymous mutations over non-synonymous ones, this means that the phylogenetic signal disappears much more rapidly from DNA sequences than from the encoded proteins. It is therefore preferable to align coding DNA at the amino acid level and it is for this purpose we have constructed the program RevTrans. RevTrans constructs a multiple DNA alignment by: (i) translating the DNA; (ii) aligning the resulting peptide sequences; and (iii) building a multiple DNA alignment by ‘reverse translation’ of the aligned protein sequences. In the resulting DNA alignment, gaps occur in groups of three corresponding to entire codons, and analogous codon positions are therefore always lined up. These features are useful when constructing multiple DNA alignments for phylogenetic analysis. RevTrans also accepts user-provided protein alignments for greater control of the alignment process. The RevTrans web server is freely available at http://www.cbs.dtu.dk/services/RevTrans/.
DNA- VERSUS PROTEIN-ALIGNMENTS
Alphabet size
The small size of the DNA ‘alphabet’ makes alignment of nucleotide sequences inherently difficult: even a pair of completely unrelated DNA sequences will typically display ∼25% identity over their entire length and it is often possible to find extended local alignments where >50% of the aligned nucleotides are identical. This makes the task of distinguishing true homology from random similarity difficult and the phylogenetic signal therefore very quickly disappears as DNA sequences diverge (1). In contrast, the simple fact that there are 20 different amino acids means that the ‘signal-to-noise ratio’ in protein–protein alignment is much better.
Silent mutations
Due to the degeneracy of the genetic code, not all mutations result in amino acid changes. Such ‘silent mutations’ typically have a small impact on organismal fitness and are therefore rarely selected against. Amino acid changing mutations, on the other hand, may negatively affect protein function and will therefore frequently be removed by purifying selection. For these reasons, a DNA sequence typically evolves more rapidly than the protein sequence it encodes (1,2). In fact the degeneracy of the genetic code means that it is theoretically possible for two very similar proteins to be encoded by a pair of DNA sequences that share only limited similarity.
Substitution matrices
Besides the information-theoretical and code-related advantages mentioned above, protein sequence alignment also benefits from the fact that most amino acid replacements are conservative in terms of physico-chemical properties. This ‘prior knowledge’ about protein evolution is captured in substitution matrices such as BLOSUM62 (3). These matrices contain empirically derived scores for each possible amino acid pair and provide a rational basis for aligning non-identical amino acids. Empirical matrices also account for unequal amino acid frequencies: if a rare amino acid is aligned with itself, then this yields a higher score than when aligning more frequently occurring residues. The fact that the overall pattern of amino acid substitution is fairly similar across protein families, means that these empirical scoring matrices can be applied to a wide range of protein alignment and database search problems. There are, of course, similar matrices for DNA. However, the pattern of nucleotide substitution is highly variable between different genes and organisms, making it difficult to construct generally applicable matrices that contain individual scores for all possible nucleotide pairs. Instead, scoring matrices for DNA typically distinguish between only two different kinds of substitution (transition and transversion), and furthermore assign the same score to all four identities (1).
ALIGNMENT OF CODING DNA
Combining information from amino acid and DNA sequences
Because of the above-mentioned properties, it is clearly preferable to take the amino acid level into account during alignment of protein-encoding DNA (4,5). By aligning coding DNA at the DNA level, one is, in effect, ignoring the information present in the genetic code and also the prior knowledge represented by amino acid substitution matrices. It should be noted that for the same reasons one should never use the DNA sequence of a protein-encoding gene for searching a database: sensitivity is much higher when the amino acid sequence is used instead (1,6,7).
Returning to alignment of coding DNA, the information at the protein level can be included by the following three-step procedure: (i) virtual translation of the coding DNA; (ii) alignment of the resulting amino acid sequences; and (iii) construction of the DNA alignment by using the multiply aligned protein sequences as a scaffold. We have constructed the program RevTrans (‘Reverse Translation of protein alignments’) to perform this procedure. The RevTrans server takes as its input a set of unaligned DNA sequences. It then translates the DNA, constructs a multiple alignment of the resulting peptide sequences, and finally builds a multiple DNA alignment by ‘reverse translation’ of the amino acid alignment. The RevTrans server will also accept user-provided protein alignments, allowing the user more control of the alignment process. For this purpose RevTrans has a number of different approaches for determining how the DNA and protein sequences map onto each other.
Alignment gaps and codon boundaries
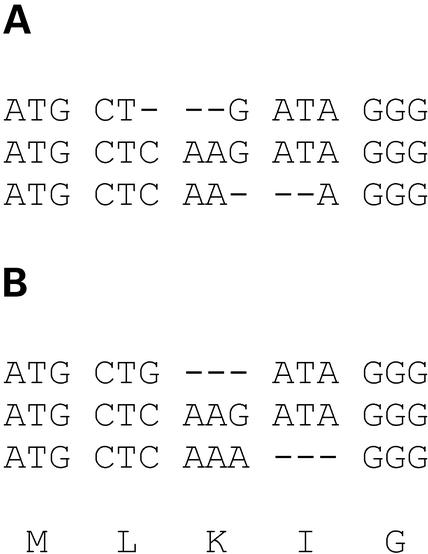
Note that a single gap in the protein alignment corresponds to a group of three gaps in the DNA alignment. This means that the DNA will be aligned in a manner that respects codon–codon boundaries (Fig. 1) and analogous codon positions will therefore always line up. For instance, a nucleotide that is located at position 3 in a given codon, will always be aligned to other 3rd-codon-position nucleotides in homologous codons (or, alternatively, to gaps). Respect of codon-boundaries and proper alignment of analogous codon positions is by no means guaranteed in simple multiple DNA alignments.
Figure 1.
Multiple alignment of coding DNA. (A) How alignment at the DNA level may lead to incorrectly aligned codon–codon boundaries. (B) How alignment of coding DNA at the amino acid level yields an alignment where analogous codon positions are properly lined up. The encoded amino acids are indicated at the bottom of (B).
By constructing the multiple DNA alignment in this way, we are in effect assuming a model of evolution where nucleotide insertions and deletions (in coding DNA) always occur in multiples of three and always start and stop at codon-boundaries. Due to the deleterious effect of frame shift mutations it is probably reasonable to assume that indels are a multiple of three nucleotides long in the vast majority of cases. There are, of course, cases where two balanced frame shift mutations have occurred near each other or where a frame shift has occurred near the stop codon but these are presumably rare. The requirement that indels always have to start and stop at codon boundaries is less likely to reflect biological reality, but has been accepted here to keep the computational burden low.
The program GenAl (5,8) is a useful alternative for constructing pairwise (not multiple) alignments of coding DNA without the above-mentioned constraints. GenAl performs pairwise alignment of DNA sequences, while taking amino acid information into account and at the same time allowing frame shift mutations and indels that can start and stop at any position. The program COMBAT (9) addresses the same problem in a slightly different manner, but again for pairwise alignment only. The server http://bioweb.pasteur.fr/seqanal/interfaces/protal2dna.html has a functionality that is very similar to RevTrans, although without automatic translation and protein alignment.
USES FOR RevTrans
RevTrans is useful in cases where a multiple alignment of coding DNA forms the basis for further investigations. This is often the case in phylogenetic analysis, where a multiple alignment is interpreted as a statement of homology with each column representing characters of common descent. In this context, proper alignment of codon boundaries is especially important for analyses that involve estimation of the ratio between non-synonymous and synonymous mutation rates (10,11).
Another use of RevTrans is as an aid for designing degenerated PCR primers. A scenario for this could be designing PCR primers targeted against a specific gene across a range of organisms. The traditional way of doing this is by aligning peptide sequences from all the organisms, identifying suitable regions for primer targeting and then designing primers that are degenerated with regards to the amino acids in the target area. By using knowledge of the actual codons used in the target area, it is possible to limit the degree to which the primers need to be degenerated. RevTrans makes such an analysis easy to perform, and is especially useful if the chosen target area aligns poorly in a DNA alignment.
‘USER MANUAL’
The RevTrans web server is freely available at http://www.cbs.dtu.dk/services/RevTrans/ and the command line version of the program can be downloaded from the same URL.
The web interface to RevTrans aims at being easy to use and intuitive. Sequence data can be entered by uploading files from the users hard disk or by pasting directly into the browser window. If the user only submits DNA data, then RevTrans automatically translates them into amino acid sequences (using the standard genetic code), invokes the dialign2 program (12) to construct a multiple alignment of these protein sequences and finally constructs the DNA alignment that is then presented on a web page together with possible errors or warnings. Both the resulting DNA alignment and the intermediate peptide alignment can be downloaded as text files.
The RevTrans server also accepts user-provided peptide-alignments to allow users greater control of the protein alignment step: if the user submits a protein alignment in addition to the DNA sequences, then RevTrans automatically uses that as a scaffold for constructing the DNA alignment. RevTrans also allows users to simply translate their DNA sequences without simultaneously aligning them and without getting a DNA alignment. This is done by pressing the ‘Translate only’ button in the submission form and may be useful for users wishing to construct their own protein alignment using programs such as ClustalW/ClustalX (13,14) or DiaAlign2 (12).
RevTrans also offers a number of advanced options relating to file formats and methods for matching sequences between DNA and protein files. These options can be set directly from the main RevTrans page. The default settings should work in most cases.
File formats: RevTrans supports FASTA, MSF and ALN (Clustal) for both input and output.
- Match methods:
- ○ Translation—match corresponding DNA and peptide sequences by translation using the standard genetic code.
- ○ Name—match by name alone.
- ○ Pos—match by position in the input files.
Gap-in: gap characters in the input sequences. This is especially useful for alignments which use a mixed set of gap-indicators.
Gap-out: gap character in the resulting alignment.
Verbose: RevTrans offers debugging output at several levels of verboseness in order to assist with troubleshooting.
Acknowledgments
ACKNOWLEDGEMENTS
R.W. and A.G.P. are both supported by a grant from the Danish National Research Foundation.
REFERENCES
- 1.States D.J., Gish,W. and Altschul,S.F. (1991) Improved sensitivity of nucleic acid database searches using application-specific scoring matrices. Methods, 3, 66–70. [Google Scholar]
- 2.Yang Z. and Nielsen,R.J. (1998) Synonymous and nonsynonymous rate variation in nuclear genes of mammals. Mol. Evol., 46, 409–418. [DOI] [PubMed] [Google Scholar]
- 3.Henikoff S. and Henikoff,J.G. (1992) Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA, 89, 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hein J. (1994) An algorithm combining DNA and protein alignment. J. Theor. Biol., 167, 169–174. [DOI] [PubMed] [Google Scholar]
- 5.Hein J. and Støvlbæk,J. (1996) Combined DNA and protein alignment. Methods Enzymol., 266, 402–418. [DOI] [PubMed] [Google Scholar]
- 6.Pearson W.R. (1996) Effective protein sequence comparison. Methods Enzymol., 266, 227–258. [DOI] [PubMed] [Google Scholar]
- 7.Doolittle R.F., Feng,D.F., Johnson,M.S. and McClure,M.A. (1986) Relationships of human protein sequences to those of other organisms. Cold Spring Harb. Symp. Quant. Biol., 51, 447–455. [DOI] [PubMed] [Google Scholar]
- 8.Hein J. and Støvlbæk,J. (1994) Genomic alignment. J. Mol. Evol., 38, 310–316. [DOI] [PubMed] [Google Scholar]
- 9.Pedersen C.N.S., Lyngsø,R.B. and Hein,J. (1998) Comparison of coding DNA. Proceedings of the 9th Annual Symposium on Combinatorial Pattern Matching (CPM). Lecture Notes in Computer Science, Rutgers University, NJ. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 10.Yang Z. and Bielawski,B. (2000) Statistical methods for detecting molecular adaptation. Trends Ecol. Evol., 15, 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Z. and Nielsen,R. (2000) Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol., 17, 32–43. [DOI] [PubMed] [Google Scholar]
- 12.Morgenstern B. (1999) DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics, 15, 211–218. [DOI] [PubMed] [Google Scholar]
- 13.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 24, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]