Abstract
We describe FrameD, a program that predicts coding regions in prokaryotic and matured eukaryotic sequences. Initially targeted at gene prediction in bacterial GC rich genomes, the gene model used in FrameD also allows to predict genes in the presence of frameshifts and partially undetermined sequences which makes it also very suitable for gene prediction and frameshift correction in unfinished sequences such as EST and EST cluster sequences. Like recent eukaryotic gene prediction programs, FrameD also includes the ability to take into account protein similarity information both in its prediction and its graphical output. Its performances are evaluated on different bacterial genomes.
The web site (http://genopole.toulouse.inra.fr/bioinfo/FrameD/FD) allows direct prediction, sequence correction and translation and the ability to learn new models for new organisms.
INTRODUCTION
FrameD has been initially designed to predict genes on prokaryotic sequences that may contain frameshifts. It has been used in the framework of two GC-rich genome annotation projects (1,2). One specific property of GC-rich genomes is that most coding regions naturally induce a mirror open reading frame on the reverse strand which can induce overprediction of many overlapping genes. FrameD is based on a graph model where gene overlapping is specifically modeled which leads to a good specificity of its predictions. This model includes RBS finding, probabilistic coding models and possible protein similarities. Even if it was initially developed for GC-rich genomes, FrameD also applies to other genomes, with good performances (see Results).
FrameD specificity lies in its ability to deal with frameshifts and sequences containing arbitrary IUPAC base symbols. This makes it possible to predict and correct frameshifts not only in bacterial sequences but also in matured transcribed eukaryotic sequences such as EST, EST clusters or cDNA. FrameD has been effectively used to predict coding regions in EST clusters in (3).
Beyond the ability to perform gene and frameshift prediction, the FrameD web site offers the ability to directly build models for new organisms. A rich graphical interface allows the user not only to visualize the predicted genes and frameshifts but also all the information used to perform the prediction (coding score, RBS strength, protein similarities, GC% and GC3% local statistics).
MATERIAL AND METHODS
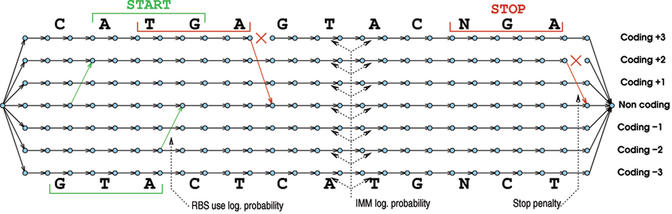
To perform gene prediction, FrameD relies on a weighted directed acyclic graph (DAG) designed in such a way that every path in the graph represents a possible gene prediction (consistent with START and STOP codons use, including possible partial genes on the sequence border). The graph in Figure 1 is an example of the graph used for a short sequence. It has seven parallel tracks that respectively correspond, from top to bottom, to the assumption that a region is coding in frame 3, 2, 1, non-coding, or coding in frame −1, −2 and −3. Each nucleotide is represented by seven edges in the graph, one on each track, represented below each nucleotide in the figure. These are called ‘content’ edges. Other edges connect vertices from the nucleotide at position i to the nucleotide at position i+1. These are called ‘signal’ edges: the occurrence of a possible translation START in a given frame p induces the creation of a signal edge that allows to go from the non-coding track to the track coding in frame p. Conversely, a STOP codon in frame p induces the creation of a signal edge that goes from the frame p coding track to the non-coding track and also prevents any path ( prediction) from crossing a STOP codon by removing the corresponding horizontal signal edge (Fig. 1). When dealing with unfinished sequences, degenerated START and STOP codons are also detected.
Figure 1.
A simplified view of the directed acyclic graph built for analyzing the sequence CATGAGTACNGA. This view ignores the additional complexity induced by gene overlapping regions and frameshift modeling. The occurrence of a START codon at position 2 to 4 induces a ‘signal’ edge that goes from the non-coding track to the +2 coding track. Similarly, the occurrence of the NGA codon at the end induces a STOP signal edge. Edge weights sources are indicated using dotted arrows.
A source-sink path in this graph represents a possible prediction. However, genes may not overlap. To allow gene overlapping, frequent in bacterial genomes, we further added six so-called ‘bi-coding’ tracks to the graph that represent regions coding in two different frames on the same strand. Each occurrence of a START codon (resp. STOP) is represented by an additional edge that allows to reach (resp. quit) the corresponding ‘bi-coding’ track.
Finally, to represent possible frameshifts, additional edges have been added that allow, for example, to reach a given coding track from another one (for a deletion, the edge jumps over one nucleotide).
In this final graph, every path from the source to the sink represents a possible prediction. In order to choose a prediction among the exponential number of paths in the graph, each edge receives a weight. If we interpret these weights as the opposite of the logarithm of the probabilities that the edge can be used in a path, a shortest path in this weighted graph represents a most reliable path (prediction).
Weighting the graph
The probability associated with content edges is defined by the emission probability of the corresponding nucleotide in a track-specific interpolated Markov model (IMM) (4). Each track is associated with a specific model (0th order Markov model for the non-coding track, three-periodic IMM for coding tracks and a mixture of two coding models for bi-coding tracks). In order to deal with noisy sequences, basic IMMs have been extended to deal with arbitrary sequences over the alphabet A,T,G,C,N and are therefore able to provide the probability of emission of a given nucleotide A,T,G,C or N after any kmer that possibly contains Ns. The basic statistics needed for the estimation of an IMM are the number of occurrences of arbitrary k+1mers (k ranging from 0 to 8 in practice) in a learning set of sequences over the alphabet A,T,G,C. For k+1mers that do not contain a degenerated N this count is obtained by enumeration of occurrences in the learning set. For k+1mers containing Ns, the number of occurrences can be computed recursively from counts associated with k+1mers containing one less N.
For signal edges, we consider that only one of all the signal edges that leave a vertex may be used, acting as a switch between tracks. The following weights are used:
A constant STOP penalty is associated with each signal edge representing a STOP occurrence.
A constant frameshift penalty is associated with each signal edge representing a frameshift (deletion and insertion are considered as equally probable).
For START codons, a RBS hybridization energy E is estimated from an approximate thermodynamic model using elementary energies (5) and a free end-gap like alignment algorithm between an RBS motif and a short region before each START codon. By analogy with thermodynamics, the probability that the START is used is then defined as α/(1+β exp(E)) where α and β are constant to be estimated.
The remaining horizontal signal edge is weighted by the logarithm of 1 minus the probability of all other signals that occur at the same position (if any, frameshift probability being considered as negligible).
When protein similarities are available, the coding score of each nucleotide is enhanced in the corresponding frame using a simple scheme. We define the mean (bit) score S of a hit as the (bit) score divided by the hit length. For each nucleotide in the sequence, the strongest mean (bit) score is used to enhance the corresponding content edge using a pseudo-count approach: if p is the original probability of the content edge for the nucleotide, the updated probability is p′=( p+γS)/(1+γS) where γ is a user-defined parameter (BlastX hits confidence). When γ is not null, the similarity information can also enhance frameshift prediction (see 6 for a related similarity-based approach to frameshift detection).
Gene and frameshift prediction
Once all parameters are fixed, the problem of finding a most reliable path in the previous directed graph can be solved using any DAG shortest path algorithm. This is done in linear time and space in the sequence length.
This algorithm will only compute one optimal prediction. In practice, it may be the case that several sub-optimal predictions differ significantly from the optimal one. This may be the case for the choice of a START codon or the use (or not) of a frameshift. To make such uncertainty in the optimal prediction visible, FrameD computes for every signal edge in the graph the probability that this edge is used over all possible predictions. This information can be interpreted as a ‘mean’ prediction that summarizes all possible predictions, taking into account their respective reliability. This computation is done using a forward-backward like dynamic programming algorithm, in linear time and space.
Fixing parameter values
In order to compute all edge weights, FrameD requires the value of several parameters.
The frameshift penalty being set to a very large value, the STOP penalty, α and β parameters have been estimated by optimizing the quality of the prediction on curated annotated sequences. The quality of the prediction was measured at the nucleotide level and at the gene level by counting correctly predicted STOP and START codons. For a given type of predicted item, sensitivity (Sn) is defined as the ratio of the number of correctly predicted items to the number of annotated items; specificity (Sp) as the ratio of the number of correctly predicted items to the number of predicted items.
The criteria optimized to estimate the parameters is the sum of these six ratios. It has been optimized using a dedicated genetic algorithm on a region of the GC-rich Sinorhizobium meliloti genome. The parameters are quite robust and provide good prediction performances on genomes with rich and medium GC content (see Results). For low GC%, another parameter set has been tuned using Rickettsia prowazekii sequences.
WEB SITE DESCRIPTION
FrameD is implemented in C++ and is available as a standalone program, downloadable from FrameD web site. The main web page allows users to specify the sequence and parameters. A precise description of the different sections in this page is available online.
The first section (Sequence) allows the user to enter a DNA sequence and either to select a probabilistic model estimated on existing bacterial/eukaryotic genomes or to build a new gene model (see below).
The ‘FrameD behavior control’ section allows to modify the default parameters used to perform gene and frameshift prediction. For bacterial sequence analysis, the user must select the GC% class (high/medium or low) for the sequence analyzed. For high/medium GC%, the internal α and β parameters used are close to the parameters obtained after optimization on S.meliloti genome. For low GC%, they have been tuned for the R.prowazekii genome. For intronless eukaryotic sequence analysis, the user must select the ‘Matured eukaryotic sequences analysis’ option which restrict start codons to ATG, deactivates the RBS search and changes a priori probabilities of being coding or not. Most importantly, the user must select a frameshift penalty that reflects the sequence quality. Other functions are available such as the translation of the predicted genes or the correction of predicted frameshifts.
The ‘Protein similarities’ section allows the user to specify existing similarities between the sequence and protein sequences. These similarities should be provided using the so-called ‘tabulated format’ available in recent versions of the NCBI-BlastX (7) program (-m8 flag in release 2.2.5). The choice of the set of similarities that are submitted to FrameD is entirely left to the user, including the choice of the expectation threshold and the protein databases used.
The ‘Output parameter’ section allows to control the layout of the prediction (textual, graphical or both, image size…). All parameters are detailed in the online help.
The results of FrameD analysis include a list of predicted CDS and frameshifts in text format followed by a graphical output that summarizes the predicted genes, frameshifts and all the information used to perform the prediction. Links to translated and frameshift corrected sequences are also available if requested.
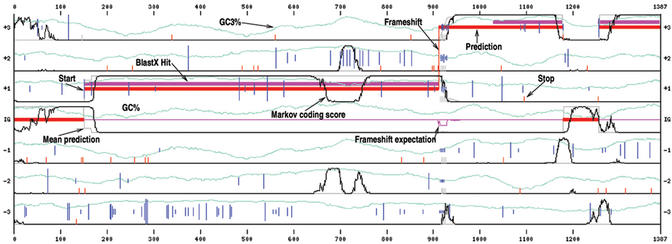
FrameD graphical output is illustrated in Figure 2 where it is applied to a genomic sequence of Ralstonia solanacearum modified to highlight FrameD features.
Figure 2.
An example of the graphical output of FrameD. The sequence is on the x-axis. The y-axis corresponds to possible predictions. From top to bottom: frame 3, 2, 1 coding tracks, intergenic track (IG) and frame −1, −2, −3 coding tracks. In-frame START codons are represented as blue vertical lines. The longer the line, the better a possible RBS. In-frame STOP codons are represented as small red vertical lines (grey if the STOP codon is degenerated). Thin black lines represent the smoothed normalized coding/non coding score. Finally, BlastX hits are represented as magenta blocks. The prediction itself is visible as red blocks and the ‘mean’ prediction as a thin grey line. The thin magenta line represents frameshift expectation. The sequence here has been specifically modified for the example: the ATG start codon of the gene, at position 148 has been replaced by an ANG and 15 nucleotides from position 915 to 929 have been replaced by 14 Ns. Using the ‘low quality sequence’ frameshift penalty, FrameD correctly predicts a gene that starts at position 148 and a frameshift between positions 911 and 912. The frameshift expectation and the ‘mean’ prediction make clear the uncertainties on the frameshift and START positions.
Another web page allows the user to build probabilistic models from user provided coding sequences. Since FrameD uses extended interpolated Markov models for building probabilistic models of coding sequences, the amount of sequence provided is not as crucial as in classical Markov models which may be prone to over-fitting. Five to ten kilobases of CDS provide a reasonable start but the more the better. Once the model is built, it is made available in the available organisms list on the main web page. A maximum of 30 such user models are stored on the site for a maximum period of 15 days.
RESULTS
We have applied FrameD and GeneMarkS (8) on six different complete genomes chosen to cover several criteria: GC percent, taxonomy, frequency of laterally transfered genes, pathogenicity. For FrameD, when it was possible, the probabilistic model was build from sequences available on SWISS-PROT before the release of the complete genome. For Pyrococcus abyssi, very few such sequences existed and we have used sequences with strong similarities with known proteins when the genome was annotated (high confidence level in the annotation). For R.solanacearum, the model used was build during the annotation of the genome. GeneMarkS builds its own models on-the-fly. Each software is used with its default parameters. No similarity information is used by FrameD. An annotated gene is considered as correctly predicted when it is predicted with the correct STOP codon.
Table 1 reports the gene level sensitivity and specificity results of this comparison. FrameD performances are comparable to the performances of GeneMarkS, with a tendency to get better sensitivity at the cost a lower specificity for genomes with a GC% above 50. We also measured the qualities of START predictions (not reported here). GeneMarkS and FrameD give very similar results but the reliability of the START position in existing annotations is probably not sufficient to conclude. Compared to GeneMarkS, which requires at least 100 kb of sequence to work, FrameD is able to analyze short sequences and offer the extra ability to reconstruct genes in the presence of frameshifts and ambiguous nucleotide symbols and therefore to process EST or EST clusters.
Table 1. Comparison of FrameD and GeneMarkS.
| Species | GC% | Type | Size (Mb) | FrameD Sn/Sp | GeneMarkS Sn/Sp |
|---|---|---|---|---|---|
| Bacillus subtilis | 43.5 | gram+ | 4.2 | 94.88/92.86 | 96.68/93.95 |
| Pyrococcus abyssi | 44.7 | archæ | 1.8 | 97.85/91.47 | 98.58/91.57 |
| Escherichia coli | 50.8 | gram− | 4.6 | 94.12/95.57 | 92.10/97.12 |
| Neisseiria meningitidis | 51.8 | gram− | 2.2 | 87.22/89.54 | 82.26/95.67 |
| Pseudomonas aeruginosa | 66.6 | gram− | 6.3 | 97.12/96.05 | 97.11/96.86 |
| Ralstonia solanacearum | 66.9 | gram− | 3.7+2.1 | 97.85/92.66 | 91.13/98.04 |
For each genome, we report its GC%, the type of the organism and the genome size. For each software, we give the gene level sensitivity (percentage of annotated genes among predicted genes) and specificity (percentage of predicted genes among annotated genes).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
REFERENCES
- 1.Salanoubat M., Genin,S., Artiguenave,F., Gouzy,J., Mangenot,S., Arlat,M., Billault,A., Brottier,P., Camus,J., Cattolico,L. et al. (2002) Genome sequence of the plant pathogen Ralstonia solanacearum. Nature, 415, 497–502. [DOI] [PubMed] [Google Scholar]
- 2.Galibert F., Finan,T.M., Long,S.R., Puhler,A., Abola,P., Ampe,F., Barloy-Hubler,F., Barnett,M.J., Becker,A., Boistard,P. et al. (2001) The composite genome of the legume symbiont Sinorhizobium meliloti. Science, 293, 668–672. [DOI] [PubMed] [Google Scholar]
- 3.Journet E.P., van Tuiner,D., Gouzy,J., Crespeau,H., Carreau,V., Farmer,M.J., Nicoel,A., Schiex,T., Jaillon,O., Chatagnier,O. et al. (2002) Exploring root symbiotic programs in the model legume Medicago truncatula using EST analysis. Nucleic Acids Res., 30, 5579–5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salzberg S.L., Delcher,A.L., Kasif,S. and White,O. (1998) Microbial gene identification using interpolated Markov models. Nucleic Acids Res., 26, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serra M., Turner,D. and Freier,S. (1995) Predicting thermodynamic properties of RNA. Methods Enzymol., 259, 243–261. [DOI] [PubMed] [Google Scholar]
- 6.Brown N.P., Sander,C. and Bork,P. (1998) Frame: detection of genomic sequencing errors. Bioinformatics, 14, 367–371. [DOI] [PubMed] [Google Scholar]
- 7.Altschul S., Madden,T., Schaffer,A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Besemer J., Lomsadze,A. and Borodovsky,M. (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes, implications for finding sequence motifs in regulatory regions. Nucleic Acids Res., 29, 2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.