Abstract
Signal search analysis is a general method to discover and characterize sequence motifs that are positionally correlated with a functional site (e.g. a transcription or translation start site). The method has played an instrumental role in the analysis of eukaryotic promoter elements. The signal search analysis server provides access to four different computer programs as well as to a large number of precompiled functional site collections. The programs offered allow: (i) the identification of non-random sequence regions under evolutionary constraint; (ii) the detection of consensus sequence-based motifs that are over- or under-represented at a particular distance from a functional site; (iii) the analysis of the positional distribution of a consensus sequence- or weight matrix-based sequence motif around a functional site; and (iv) the optimization of a weight matrix description of a locally over-represented sequence motif. These programs can be accessed at: http://www.isrec.isb-sib.ch/ssa/.
INTRODUCTION
Signal search analysis is a versatile method to detect, analyze and characterize sequence motifs that are positionally correlated with a functional site in a DNA sequence, for instance a transcription start site. The method has been developed for the analysis of eukaryotic promoters but has a much broader application potential as illustrated by the example shown in this paper. The different types of data analysis methods have been described in a series of previous papers (1–4).
Input to a signal search analysis procedure is a data structure called a functional position set. A functional position set consists of pointers to positions in DNA sequences stored in a database and is used to extract a corresponding set of fixed-length DNA sequence fragments of desired length and location relative to the functional sites. The extracted sequence fragments are then scanned in a sliding window in order to determine the local frequencies of particular sequence motifs. The size of the window and the number of bases by which it is shifted in one step are standard parameters of a signal search analysis method. Note that the window is usually several bp longer than the sequence motif so that even with a window shift of more than 1 bp no motif occurrence is missed. The sequence motifs considered are either defined by a consensus sequence (potentially including ambiguous IUPAC codes) or by a weight matrix (4). Search operations require specification of a corresponding cut-off value defined by a maximal number of allowed mismatches in the case of a consensus sequence or by a threshold score in the case of a weight matrix. Even though it is now commonly recognized that consensus sequences are inappropriate for an accurate description of a gene control signal, they are still useful to demonstrate qualitative effects in a simple and intuitively convincing manner (see example in Fig. 1). They are also useful at the discovery stage of a new sequence motif providing a first approximation which then can be used as a starting point for the development of a more accurate weight matrix-based motif definition.
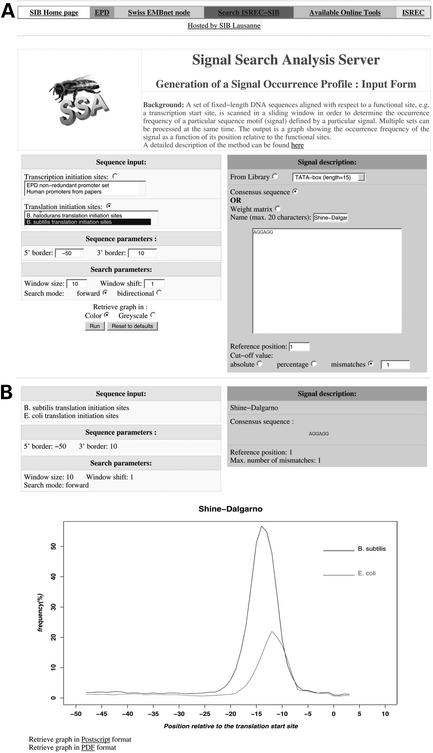
Figure 1.
(A) Input form for generating signal occurrence profiles with the program OProf. Sequences around annotated translation start sites from two bacterial genomes (left side) are searched for the hexanucleotide sequence motif AGGAGG (right side) complementary to the 3′ end of 16 s ribosomal RNA. Note that the signal is searched in a sliding window of 10 bp and that one mismatch is allowed for determining its local frequency. This analysis serves to compare the strength and location of the Shine-Dalgarno mRNA–rRNA interaction motif in Escherichia coli and Bacillus subtilis in a qualitative manner. (B) Result: note that the Shine-Dalgarno interaction motif is stronger in B.subtilis than in E.coli and centered about two bases further upstream in the former species. More than a hundred bacterial genomes are now available to perform this type of analysis.
The central concept of signal search analysis has been formalised as a ‘locally over-represented sequence motif’ (4). The definition of a locally over-represented sequence motif has three components: (i) a motif description (e.g. a consensus sequence or weight matrix); (ii) a corresponding cut-off value; and (iii) a region of preferential occurrence. For instance, the eukaryotic TATA-box is a locally over-represented sequence motif that can be approximately described by the consensus sequence TATAAA, a cut-off value of two mismatches and a region of preferential occurrence extending from 35 to 20 bp upstream of the transcription start site.
Signal search analysis has played an instrumental role in the characterization of eukaryotic promoter elements. This is one reason why we have recently decided to make these relatively old methods available via a web interface. The signal search analysis server thus serves partly educational and documentary purposes. The pages allow students and researchers to rediscover gene expression signals in an interactive manner and to confirm or challenge commonly accepted hypotheses originally derived from small sets, as illustrated by the example below. In addition and more importantly, we believe that these programs are still very useful and could enable others to discover new, or better characterize already known, control signals in the rapidly growing collection of complete genomes.
THE SIGNAL SEARCH ANALYSIS SERVER
The signal search analysis server (http://www.isrec.isb-sib.ch/ssa/) provides access to precompiled functional position sets and four different analysis programs. The functional position sets currently include collections of transcription initiations sites (promoters) from eukaryotic species and translation start sites from a large variety of prokaryotic genomes. The promoter sets are based on the Eukaryotic Promoter Database EPD (5). In the near future, we plan to offer compilations of eukaryotic pre-mRNA processing signals (splice donors, acceptors, and polyadenylation sites) as well.
The four signal search analysis programs offered by the server are:
CPr: generates a constraint profile. Input is a set of DNA sequences adjacent or around a functional site plus a ‘signal sequence collection’ composed of consensus sequence-based motif descriptions. The frequencies of these motifs are determined in a sliding window. At each position an index is computed reflecting the non-randomness of the subsequences in the corresponding window, which is then plotted as a function of the position relative to the functional site. The method was described previously (1). Examples of constraint profiles for prokaryotic and eukaryotic promoters have been shown (2).
SList: generates a signal list. The input and initial data processing steps are the same as for the generation of a constraint profile. The output consists of a list of signals that are strongly over or under-represented at a particular distance from the functional site as compared to other locations. Examples of signal lists for prokaryotic and eukaryotic promoter regions can be found (2).
OProf: generates a signal occurrence profile (Fig. 1). Input is a set of DNA sequences adjacent or around a functional site plus a sequence motif defined by a consensus sequence or a weight matrix plus a cut-off value. The frequency of the motif is determined in a sliding window. The result is a graph showing the frequency of the signal as a function of the position relative to the functional site. Examples of signal occurrence profiles for eukaryotic promoter elements can be found in the literature (3,4).
PatOp: optimizes a weight matrix description of a locally over-represented sequence motif. The inputs are the same as for the generation of a signal occurrence profile. The input sequence motif serves as an initial model for starting the optimization process. The output is a complete weight matrix-based description of a sequence motif maximizing a quantitative criterion of local over-representation, including the optimized border positions and cut-off value. The method is described and exemplified elsewhere (4). The sequence motifs of four major eukaryotic promoter elements, the TATA-, CCAAT, GC-boxes and initiator motif, were determined with this method.
EXAMPLE
In order to illustrate the method, we show an application to a prokaryotic translational control signal that is part of the ribosome-binding site. The original observation leading to the discovery of this signal was that sequences immediately upstream of prokaryotic translation start codons often contain short oligonucleotides complementary to a poly-pyrimidine-rich 3′-terminal region of 16 s ribosomal RNA (6). This control signal, which is sometimes referred to as the Shine-Dalgarno ribosome binding-site motif, was shown to increase translational efficiency of the corresponding mRNAs. In order to analyze the species-specific properties of this signal, we generated signal occurrence profiles for translation start sites of two bacterial genomes using one of the hexanucleotides complementary to the 16 s ribosomal RNA region found to interact with the leader sequences of mRNAs. The filled out input form and the results returned by the signal search server are shown in Figure 1.
Acknowledgments
ACKNOWLEDGEMENT
The signal search analysis server is partly funded by grant 31-063933.00 from the Swiss National Research Foundation.
REFERENCES
- 1.Bucher P. and Bryan,B. (1984) Signal search analysis: a new method to localize and characterize functionally important DNA sequences. Nucleic Acids Res., 12, 287–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bucher P. and Trifonov,E.N. (1986) Compilation and analysis of eukaryotic POL II promoter sequences. Nucleic Acids Res., 14, 10009–10026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bucher P. and Trifonov,E.N. (1988) CCAAT-box revisited: bidirectionality, location and context. J. Biomol. Struct. Dyn., 5, 1231–1236. [DOI] [PubMed] [Google Scholar]
- 4.Bucher P. (1990) Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol., 212, 563–578. [DOI] [PubMed] [Google Scholar]
- 5.Praz V., Perier,R., Bonnard,C. and Bucher,P. (2002) The Eukaryotic Promoter Database, EPD: new entry types and links to gene expression data. Nucleic Acids Res., 30, 322–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shine J. and Dalgarno,L. (1975) Determinant of cistron-specificity in bacterial ribosomes. Nature, 254, 34–38. [DOI] [PubMed] [Google Scholar]