Abstract
INCLUSive is a suite of algorithms and tools for the analysis of gene expression data and the discovery of cis-regulatory sequence elements. The tools allow normalization, filtering and clustering of microarray data, functional scoring of gene clusters, sequence retrieval, and detection of known and unknown regulatory elements using probabilistic sequence models and Gibbs sampling. All tools are available via different web pages and as web services. The web pages are connected and integrated to reflect a methodology and facilitate complex analysis using different tools. The web services can be invoked using standard SOAP messaging. Example clients are available for download to invoke the services from a remote computer or to be integrated with other applications. All services are catalogued and described in a web service registry. The INCLUSive web portal is available for academic purposes at http://www.esat.kuleuven.ac.be/inclusive.
INTRODUCTION
Unraveling transcriptional regulation from microarray data raises a 2-fold challenge. The first challenge is to cluster genes into biologically meaningful groups; the second is to find similar regulatory motifs (mostly transcription factor binding sites) in the promoter regions of the genes in such groups, the latter hopefully explaining the former (1).
The INCLUSive portal meets these challenges by covering the following techniques:
Normalization of microarray data.
Grouping of genes through clustering of microarray data.
Refinement and validation of the groups using Gene Ontology.
Algorithms for the detection of known and unknown regulatory motifs in the upstream sequences of the genes in the groups.
As compared to the previous release of INCLUSive (2), substantial improvements have been made. Modules for normalization of microarray data and refinement and validation of clusters have been added and the existing modules underwent a reconstruction to improve and broaden functionality. We promote a more organism-oriented approach to improve retrieval of intergenic sequences and functional information.
Besides, a web service environment was added to address the lack of interoperability among data and service providers in bioinformatics (3). All tools are provided as web services and are registered in a service registry for easy discovery and binding by remote applications. This is the first time that a suite of web services for microarray and regulatory sequence analysis is available to the bioinformatics community.
As compared to Expression Profiler (http://ep.ebi.ac.uk/), the major advantage of INCLUSive is its loosely coupled structure. All tools can be used separately as well as in a complex sequence of analysis steps. The web service architecture allows easy integration of new or alternative tools, which makes the system dynamic and flexible.
THE INCLUSive PORTAL
This section gives an overview of the different tools and algorithms available via the portal. Most of them can be used in three different ways:
By using the forms on the different web pages.
By invoking their web services from a remote computer.
By downloading and installing the stand-alone versions of the algorithms.
The web pages allow the user to upload data files and to specify the organism under study. At this moment, the analysis of data from yeast, Arabidopsis, mouse and human is fully supported. The data files have to contain data values for each gene (e.g. raw microarray intensities), combined with a unique identifier for each gene. The system supports GenBank and Unigene accession numbers, as well as yeast identifiers.
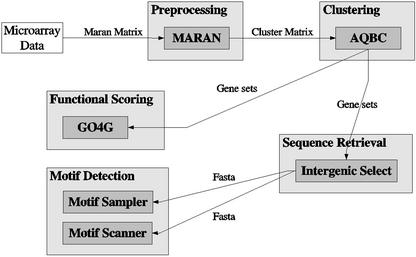
Figure 1 shows the flowchart of the INCLUSive portal. It schematizes how all modules are connected and visualizes the data flow between them.
Figure 1.
Schematic overview of the data flow between the different modules of INCLUSive. The flow supports complex analysis of microarray data, comprising ANOVA normalization, filtering and clustering, functional scoring of gene clusters, sequence retrieval, and detection of known and unknown regulatory elements. All modules are independent of each other an can be used separately. The INCLUSive web portal is available for academic purposes at http://www.esat.kuleuven.ac.be/inclusive.
For every tool, a short description is given, along with the methods used or a reference describing them.
Maran: normalizing microarray data
Maran normalizes microarray data by constructing a generic ANOVA model based upon several sources of variation in the experiment (4). The residuals obtained from fitting the model can be used for statistical inference. Other features of the Maran web application are a Loess fit procedure and an option for detecting genes that have a significantly changing expression profile.
Adaptive quality-based clustering of microarray data
The adaptive quality-based clustering (AQBC) method clusters microarray data in an heuristic iterative two-step process (5). One of the characteristics of the algorithm is that all clusters have a constant quality, represented by the significance level S. The default value for S guarantees that a gene has a probability of 95% to truly belong to the cluster it is assigned to, according to a probabilistic model of the data. As a consequence, the clusters found by AQBC contain few false positives and are thus ideal seeds for further cis-regulatory analysis.
Information Select: retrieving additional information of clustered genes
Central to Information Select is a series of organism-specific knowledge bases. At the moment, they mainly contain mappings of different public database identifiers. These knowledge bases allow the Information Select algorithm to fully characterize the genes of each cluster by providing links to a myriad of different public databases, starting from GenBank or Unigene accession numbers. Based on the organism that is specified by the user, the algorithm addresses the correct knowledge base and fetches and returns links to additional information sources, such as LocusLink, Ensembl, GeneCards, MGI, SWISS-PROT, and so on. This way, the user is spared the burden of retrieving all this relevant background information manually. In most cases, the user wants to combine information from different sources, which can be a very cumbersome and time-consuming task if done manually.
Functional scoring with GO4G
GO4G is designed to assign general functional trends to groups of genes. The algorithm extracts Gene Ontology (GO) terms (6) associated to a group of genes (a cluster for example) and calculates which terms are statistically over-represented when compared to their expected frequencies.
Used method: GO4G retrieves all annotated GO terms of a group of genes. For each term it recursively adds all parents up to the root of the tree. All terms are counted and their frequencies are compared to their expected frequencies. The latter are obtained from all annotated genes of a certain genome. A term is over-represented if its frequency differs with statistical significance from its expected frequency. For every term a p-value is calculated using a hypergeometric distribution.
Intergenic Select: retrieving intergenic sequences
Since sequence retrieval for organisms with fully sequenced genomes is well supported by other systems, we chose to cross-reference to their functionalities. You can use either Toucan (7) or EnsMart (http://www.ensembl.org/EnsMart) (8) for the selection of upstream/intergenic sequences. Both systems allow the user to select exactly the genomic regions of interest.
The alternative Intergenic Select service we provide within INCLUSive is based on an iterative BLAST-search against GenBank (NCBI). The tool accepts accession numbers and gene names to retrieve seed sequences. The BLAST hits of these sequences are used in the subsequent steps, until the required length of sequence upstream of the coding region is reached. This approach has been proven to be useful when working with compact genomes (9,10).
MotifSampler: finding over-represented motifs
The MotifSampler is a user-friendly Gibbs sampling implementation that allows detection of statistically over-represented motifs in a set of unaligned sequences (9–11). The algorithm determines in which sequences and at what positions a statistically over-represented motif is present, compared to a background model derived from the input data or compared to a user-specified organism-dependent background model.
MotifScanner: screening for known motifs
The MotifScanner is designed to search for putative sites of known motifs from TRANSFAC (12) and PlantCARE (13) in a set of sequences (7). The motifs, represented by a position probability matrix, are assumed to be hidden in a noisy background sequence, represented by a higher-order Markov model. The algorithm is based upon the core modules of the MotifSampler.
Toucan: workbench for regulatory sequence analysis on metazoan genomes
Toucan (7) is a stand-alone application for the detection of cis-regulatory elements in promoter regions of higher eukaryotes. It uses and integrates several INCLUSive modules and shows clearly the advantages of working with web services. Whatever version of Toucan is used, the invoked service always runs the latest version of the algorithm. The processor usage of the computer running Toucan is kept low because the heavy calculations are performed remotely on our Linux cluster. Also, the total file size of the application is kept low, which improves download times.
FUTURE DEVELOPMENTS
Towards future releases we plan to provide specific support for more organisms with the focus on prokaryotes. In the short term, we are preparing the deployment of services for:
Text mining-based tools for functional validation of gene clusters (14).
Detection of motif modules in sets of coregulated genes in higher eukaryotes.
In the long term we plan to add services for:
Upload of MAGE-ML files to the system (15).
Alternative clustering and meta-clustering.
Feature extraction, classification and detection of differential expression.
Comparative genome analysis and phylogenetic footprinting.
Prediction of promoter regions in eukaryotic genomes.
Inference of genetic networks.
Besides, information parsers will be added to the Information Select system. The parsed information can be used for supervised clustering, to prepare kernel documents for text mining applications and as prior knowledge for genetic network inference.
Acknowledgments
ACKNOWLEDGEMENTS
B.C., S.A., F.D.S. and P.G. are research assistants of the KU Leuven. G.T. and K.E. are research assisants of IWT. J.M. is a post-doctoral researcher of the KU Leuven. K.M. and Y.M. are post-doctoral researchers of FWO-Vlaanderen and assistant professors at the KU Leuven. B.D.M. is a full professor at the KU Leuven. Research supported by Research Council KUL: [GOA-Mefisto 666, IDO (IOTA Oncology, Genetic networks), several PhD/postdoc and fellow grants]; Flemish Government: [FWO: PhD/postdoc grants, projects G.0115.01 (microarrays/oncology), G.0240.99 (multilinear algebra), G.0407.02 (support vector machines), G.0413.03 (inference in bioi), G.0388.03 (microarrays for clinical use), G.0229.03 (ontologies in bioi), research communities (ICCoS, ANMMM)]; AWI: (Bil. Int. Collaboration Hungary/Poland); IWT: [PhD Grants, STWW-Genprom (gene promotor prediction), GBOU-McKnow (Knowledge management algorithms), GBOU-SQUAD (quorum sensing), GBOU-ANA (biosensors)]; Belgian Federal Government: [DWTC: IUAP IV-02 (1996–2001) and IUAP V-22 (2002–2006)]; EU: (CAGE); ERNSI; Contract Research/agreements: (Data4s, Electrabel, Elia, LMS, IPCOS, VIB).
REFERENCES
- 1.Moreau Y., De Smet,F., Thijs,G., Marchal,K. and De Moor,B. (2002) Functional bioinformatics of microarray data: from expression to regulation. Proceedings of the IEEE, 90, 1722–1743. [Google Scholar]
- 2.Thijs G., Moreau,Y., De Smet,F., Mathys,J., Lescot,M., Rombauts,S., Rouzé,P., De Moor,B. and Marchal,K. (2002) INCLUSive: INtegrated CLustering, Upstream sequence retrieval and motif Sampling. Bioinformatics, 18, 331–332. [DOI] [PubMed] [Google Scholar]
- 3.Stein L. (2002) Creating a bioinformatics nation. Nature, 417, 119–120. [DOI] [PubMed] [Google Scholar]
- 4.Engelen K., Coessens,B., Marchal,K. and De Moor,B. (2003) MARAN: normalizing micro-array data, Bioinformatics, 19, 893–894. [DOI] [PubMed] [Google Scholar]
- 5.De Smet F., Mathys,J., Marchal,K., Thijs,G., De Moor,B. and Moreau,Y. (2002) Adaptive quality-based clustering of gene expression profiles. Bioinformatics, 18, 735–746. [DOI] [PubMed] [Google Scholar]
- 6.The Gene Ontology Consortium (2000) Gene Ontology: tool for the unification of biology. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aerts S., Thijs,G., Coessens,B., Staes,M., Moreau,Y. and De Moor,B. (2003) TOUCAN: deciphering the cis-regulatory logic of coregulated genes. Nucleic Acids Res., 31, 1753–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clamp M., Andrews,D., Barker,D., Bevan,P., Cameron,G., Chen,Y., Clark,L., Cox,T., Cuff,J., Curwen,V. et al. (2003) Ensembl 2002: accommodating comparative genomics. Nucleic Acids Res., 31, 38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thijs G., Marchal,K., Lescot,M., Rombauts,S., De Moor,B., Rouzé,P. and Moreau,Y. (2002) A Gibbs Sampling method to detect over-represented motifs in upstream regions of coexpressed genes. J. Comput. Biol., 9, 447–464. [DOI] [PubMed] [Google Scholar]
- 10.Marchal K., Thijs,G., De Keersmaecker,S., Monsieurs,P., De Moor,B. and Vanderleyden,J. (2003) Genome-specific higher-order background models to improve motif detection. Trends Microbiol., 11, 61–66. [DOI] [PubMed] [Google Scholar]
- 11.Thijs G., Lescot,M., Marchal,K., Rombauts,S., De Moor,B., Rouzé,P. and Moreau,Y. (2001) A higher-order background model improves the detection of promoter regulatory elements by Gibbs sampling. Bioinformatics, 17, 1113–1122. [DOI] [PubMed] [Google Scholar]
- 12.Wingender E., Chen,X., Hehl,R., Karas,H., Liebich,I., Matys,V., Meinhardt,T., Prüß,M., Reuter,I. and Schacherer,F. (2000) TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res., 28, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lescot M., Déhais,P., Thijs,G., Marchal,M., Moreau,Y., Van de Peer,Y., Rouzé,P. and Rombauts,S. (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res., 30, 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glenisson P., Antal,P., Mathys,J., Moreau,Y., and De Moor,B. (2003) Evaluation of the vector space representation for text-based gene clustering. Proc. Eighth Annu. Pac. Symp. Biocomp., 8, 391–402. [DOI] [PubMed] [Google Scholar]
- 15.Spellman P.T., Miller,M., Stewart,J., Troup,C., Sarkans,U., Chervitz,S., Bernhart,D., Sherlock,G., Ball,C., Lepage,M. et al. (2002) Design and implementation of microarray gene expression markup language (MAGE-ML). Genome Biol., 3, 0046.1–0046.9. [DOI] [PMC free article] [PubMed] [Google Scholar]