Abstract
A fundamental challenge facing biologists is to identify DNA binding sites for unknown regulatory factors, given a collection of genes believed to be coregulated. The program YMF identifies good candidates for such binding sites by searching for statistically overrepresented motifs. More specifically, YMF enumerates all motifs in the search space and is guaranteed to produce those motifs with greatest z-scores. This note describes the YMF web software, available at http://bio.cs.washington.edu/software.html.
DESCRIPTION
One of the current challenges facing biologists is the discovery of novel nucleic acid binding sites for unknown regulatory factors, given a collection of genes believed to be coregulated. There are many possible sources for such putatively coregulated genes, including expression microarray experiments, gene knockout experiments and functional classes from the literature.
A number of algorithms have been proposed for the discovery of motifs in biological sequences. Many of these algorithms are designed to find more general motifs than are required for identifying transcription factor binding sites. These algorithms are based on methods such as expectation maximization (1), Gibbs sampling (2,3) and greedy algorithms (4), any of which may terminate in a locally optimal solution.
The number of well conserved bases in the collection of binding sites of a single transcription factor is frequently small enough that, for this particular problem, one need not rely on such general local search heuristics. Instead, one can afford to use enumerative methods that guarantee global optimality. This is the approach we described in earlier work (5,6). The algorithm described in that work is called YMF (Yeast Motif Finder, as the motif model was derived from a study of known transcription factor binding sites in the yeast Saccharomyces cerevisiae), an enumerative algorithm that, given the regulatory regions of several related genes, is guaranteed to produce the motifs with greatest z-scores. The z-score of a motif is the number of standard deviations by which its observed number of instances in the actual input sequences exceeds its expected number of instances, had the input sequences instead been random. The motifs themselves are short sequences over the IUPAC alphabet, with N's (called ‘spacers’ in what follows) constrained to occur in the middle of the sequence.
In previous work (5), we discussed the results of validation experiments in which YMF was used to identify candidate binding sites in 23 well studied regulons of S.cerevisiae. For 18 of these regulons YMF succeeded in reporting the known binding site consensus for the regulon's principal transcription factor. We then turned to gene families in the functional and mutant phenotype catalogues of S.cerevisiae from the MIPS database, where YMF reported many promising novel transcription factor binding sites. Park et al. (7) used YMF to predict the binding sites of the transcription factor DosR in the Mycobacterium tuberculosis genome.
YMF is available at http://bio.cs.washington.edu/software.html both in source code and through a web interface. This note describes the web interface to YMF. The reader is referred to earlier work (5,6) for details on YMF's algorithm and its application to promoter sequences.
Although users may appreciate guidance concerning which motif discovery algorithm to choose, there are no accepted criteria to predict when one such algorithm will be more successful than another. We have performed experiments (8) on both synthetic and S.cerevisiae promoter sequences, comparing YMF to MEME (1) and AlignACE (3), which are popular tools used for the discovery of regulatory elements. In those experiments YMF was found to make more accurate predictions of the known regulatory elements on more of the S.cerevisiae regulons than the other tools. However, one of the conclusions derived from those experiments is that it may be beneficial to try a few very different motif discovery tools in addition to YMF, as different data sets seem to yield to different tools.
USER INPUTS
The simple web form asks the user to supply the regulatory input sequences in Fasta format. The user also chooses a few parameter values that specify the type of motif YMF should report: the motif size (not including any spacers—N's—that YMF will insert), the maximum number of spacers YMF should insert, and the maximum number of other degenerate IUPAC symbols YMF should employ. For instance, the motif TCRNNNACG has motif size 6, with three spacers and one degenerate symbol R. The smaller each of these chosen values is, the faster YMF will run.
Finally, the user must specify the organism from which the input sequences were collected. The reason is that YMF reports motifs that are statistically overrepresented in the input sequences with respect to a ‘background’ model that captures the promoter regions of all genes from the organism. YMF has precomputed these background models for several common genomes. If the user's genome is not among these, there is a simple auxiliary web form that asks for a Fasta file containing all the promoter regions for that genome. Once YMF has processed these to create the background model, it will be available in a pull-down menu of user-created organisms for the current and future YMF sessions.
INITIAL YMF RESULTS
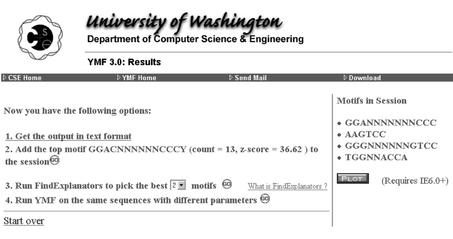
Figure 1 shows an example of the Results page that YMF produces when it has completed its enumeration and statistical analysis of all possible motifs that fit the user's parameter values. This page shows the motif (GGACNNNNNNCCCY) with the highest z-score (36.62), together with the number of instances of that motif in the input sequences (13). YMF counts instances on both DNA strands, and allows for any number of instances per input sequence.
Figure 1.
Initial ‘Results’ page of YMF.
The user now has several choices of how to proceed, as shown in Figure 1:
The top motif reported can be added to the current session.
The motif occurrences in the current session can be plotted, as shown in Figure 2.
The user can inspect the entire YMF output, which is a long list of the top motifs and their number of occurrences in the input sequences, sorted by z-score.
FindExplanators can be run on the entire YMF output, in order to select the best few motifs whose occurrences are independent of each other. See Blanchette and Sinha (9) for details.
YMF can be restarted on the same sequences with new parameter values.
YMF can be restarted with a clean session.
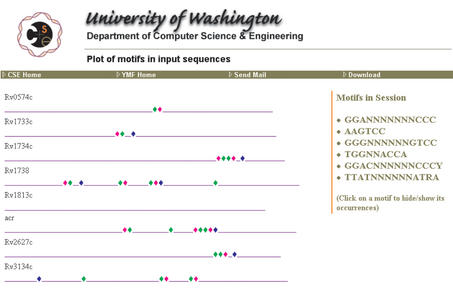
Figure 2.
YMF plot of the upstream regions of eight M.tuberculosis genes regulated by the transcription factor dosR, from Park et al. (7). The motifs presently plotted are GGANNNNNNNCCC (green), AAGTCC (blue), and GGACNNNNNNCCCY (magenta).
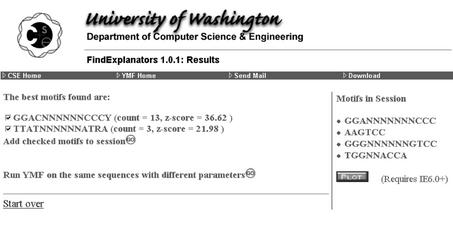
Figure 3 shows the results of running FindExplanators to choose the two best independent motifs, GGACNNNNNNCCCY and TTATNNNNNNATRA. Note that the first of these is the same as the best motif reported by YMF in Figure 1, which will always be the case. Most of the user options listed above are again available to the user at this point.
Figure 3.
Results of running FindExplanators.
PLOTTING THE MOTIFS
Figure 2 shows the result of plotting, once the six motifs listed at the right side of that figure have been added to the session. Each horizontal line is a schematic representing one of the input sequences. YMF labels each line with the name provided by the user in the corresponding Fasta annotation line in the input sequence file. The colored diamonds along each line indicate the positions of motif instances, with each motif in the figure assigned a unique color. By clicking on a motif at the right side of the screen, the user prompts YMF to show or hide the instances of that motif. In Figure 2, all instances of three of the six session motifs are currently being shown.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the referees for careful testing of the web interface and for making several good suggestions for improvements. This material is based upon work supported in part by a Microsoft fellowship, in part by the National Science Foundation under grants DBI-9974498 and DBI-0218798 and in part by the National Institutes of Health under grant HG02602-01.
REFERENCES
- 1.Bailey T.L. and Elkan,C. (1995) Unsupervised learning of multiple motifs in biopolymers using expectation maximization. Machine Learning, 21, 51–80. [Google Scholar]
- 2.Lawrence C.E., Altschul,S.F., Boguski,M.S., Liu,J.S., Neuwald,A.F. and Wootton,J.C. (1993) Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science, 262, 208–214. [DOI] [PubMed] [Google Scholar]
- 3.Roth F.P., Hughes,J.D., Estep,P.W. and Church,G.M. (1998) Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat. Biotechnol., 16, 939–945. [DOI] [PubMed] [Google Scholar]
- 4.Hertz G.Z. and Stormo,G.D. (1999) Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics, 15, 563–577. [DOI] [PubMed] [Google Scholar]
- 5.Sinha S. and Tompa,M. (2002) Discovery of novel transcription factor binding sites by statistical overrepresentation. Nucleic Acids Res., 30, 5549–5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinha S. and Tompa,M. (2000) A statistical method for finding transcription factor binding sites. In Bourne,P., Gribskov,M., Altman,R., Jensen,N., Hope,D., Lengauer,T., Mitchell,J., Scheeff,E., Smith,C., Strarde,S., and Weissig,H. (eds), Proceedings of the Eighth International Conference on Intelligent Systems for Molecular Biology, AAAI Press, Menlo Park, CA, pp. 344–354. [PubMed] [Google Scholar]
- 7.Park H.-D., Guinn,K., Harrell,M.I., Liao,R., Voskull,M.I., Tompa,M., Schoolnik,G.K. and Sherman,D.R. (2003) Rv3133c/dosR is a transcription factor that mediates the hypoxic response of M. tuberculosis. Mol. Microbiol., 48, 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinha S. and Tompa,M. (2003) Performance comparison of algorithms for finding transcription factor binding sites. In Third IEEE Symposium on Bioinformatics and Bioengineering, IEEE Press, Los Alamitos, pp. 214–220. [Google Scholar]
- 9.Blanchette M. and Sinha,S. (2001) Separating real motifs from their artifacts. Bioinformatics, 17, S30–S38. [DOI] [PubMed] [Google Scholar]