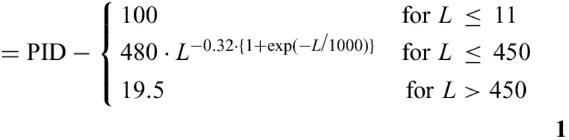Abstract
UniqueProt is a practical and easy to use web service designed to create representative, unbiased data sets of protein sequences. The largest possible representative sets are found through a simple greedy algorithm using the HSSP-value to establish sequence similarity. UniqueProt is not a real clustering program in the sense that the ‘representatives’ are not at the centres of well-defined clusters since the definition of such clusters is problem-specific. Overall, UniqueProt is a reasonable fast solution for bias in data sets. The service is accessible at http://cubic.bioc.columbia.edu/services/uniqueprot; a command-line version for Linux is downloadable from this web site.
INTRODUCTION
The problem of biased data sets
Increasingly often experimentalists face the problem of searching for some ‘significant’ motifs or features in a set of proteins retrieved from common database searches. When we simply use the sequences with today's bias, we risk to over-estimate significance (1). The bias has two potential sources: (i) certain families could be missing; or (ii) could be over-represented. Such bias may hinder finding sequence-patterns that are related to protein structure and/or function. We cannot solve the first problem since we do not have any insight into the still undiscovered and missing sequences of the protein universe. However, we can discard over-represented sequences by grouping similar proteins.
Inferring functional similarity from sequence similarity
Supposedly, the mostly desired criterion for grouping two proteins into one ‘family’ is that the two share a common function. This is by far not an easy task considering the many different levels of functional roles any particular protein orchestrates within a living cell. In fact, while such inferences are accurate for high levels of pairwise sequence similarity, they become accurate rather rapidly with the level of divergence between the two proteins (1,2). If we consider two proteins to have similar function by the token that both participate in cell cycle control, we need to establish different thresholds for pairwise sequence similarity that allows to infer this feature by homology (K.O.Wrzeszczynski and B.Rost, manuscript submitted). We need to apply yet a different battery of thresholds to infer that: (i) two proteins dwell in the same sub-cellular compartment (3, K.O.Wrzeszczynski and B.Rost, manuscript submitted); (ii) that they belong to the same groups of cellular function (4), have similar binding sites (5) or belong to similar descriptions according to the GeneOntology (6,7).
Inferring structural similarity from sequence similarity
Arguably, the feature that is most conserved with evolutionarily diverging sequences is protein structure (8–10). If we consider protein sequences as simple strings of letters, mathematics suggests that the probability of finding 10 in 20 aligned residues (50%) is much higher than that of finding 100 in 200 (also 50%) (11). Sander and Schneider (12) accounted for this obvious reality of sequence analysis by introducing an empirical threshold that related alignment length and pairwise sequence identity in a way allowing to automatically determine families of proteins with similar structure in their HSSP database. A refined version of this original HSSP curve proved to better discriminate between proteins of similar and non-similar structure than expectation values from pairwise BLAST searches (9). Since the functional form of this curve also appears to rather accurately reflect similarity in sub-cellular localisation (3, K.O.Wrzeszczynski and B.Rost, manuscripts submitted) and enzymatic activity (1), we based our bias-reduction tool UniqueProt on this curve. UniqueProt removes the bias of sequence-redundant proteins from a given data set in the hope of acquiring unique sub-sets that constitute more accurate approximations to the goal of analysing sets representative for the protein universe. However, users should be careful about submitting data sets with very heterogeneous domain architectures since the UniqueProt algorithm may completely remove domain-representatives. Especially the submission of sequence-fragments is not recommended.
METHOD
Input
The program accepts either a set of sequences in FASTA format or a list of identifiers from either of the following protein databases: SWISS-PROT (13), PDB (14) or TrEMBL (13). Alternatively, one of the following alignment-file formats is accepted to bypass the first step of the algorithm (see below): BLAST, PSIBLAST, pair, markx0, markx1, markx2, markx3, markx10 or srspair.
HSSP-value to measure sequence similarity
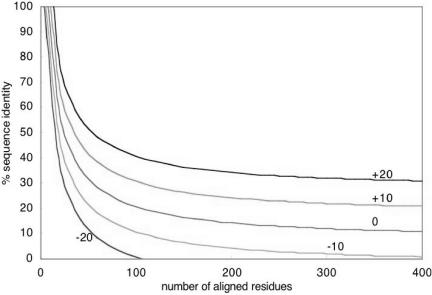
First all sequences are compared with BLAST (15,16). Then the percentage of identical residues and the length (L) of the BLAST-derived alignment (without including the gaps) are converted into the HSSP-value (HV) according to Eq. 1. Here PID is the number of identical residues in the BLAST alignment times 100 and divided by L. The HSSP-value reflects whether an alignment is above the HSSP-curve (9,12) (HSSP-value >0) or below (<0) (Fig. 1). For the first case (>0) the HSSP-value can be seen as a degree of sequence-proximity whereas for the latter case (<0) it gives an estimate about the distance between two compared sequences. For the case that an alignment file instead of a FASTA file or list of identifiers is submitted the HSSP-value is directly derived from the alignment information without performing a BLAST comparison first.
Figure 1.
HSSP-curve for different values υ. The curves illustrate different HSSP-values υ from the original HSSP-curve (Eq. 1). Every pairwise alignment can be represented as a point in the graph above. Any naturally evolved two proteins for which the similarity falls above the curve υ=0 are expected to have similar structures. Higher values provide more cautious estimates about features common to two proteins and larger sequence-unique sub-sets.
Algorithm
In order to find the largest sub-set of proteins that fulfil the constraint that no pair in that set has an HSSP-value>υ (υ=user defined threshold), we applied a simple greedy algorithm similar to that employed toward this end by Hobohm and Sander (17): for each protein P in the submitted set, the algorithm counts the number of proteins NP that share an HSSP-value with P larger than υ. We consider all proteins {NP} with HV>υ as belonging to the family F(P). Next, we store the number and identifiers of all neighbours for each protein and sort the entire data set by the size of the families {F}. Finally, the greedy algorithm simply works down that list by starting at protein P′ and excluding all members of family F(P′). We start either with the largest or the smallest family (option selected by user). In particular, the algorithm is as follows. (i) Take singletons: if the family F(P′) contains only one sequence, P′ is added to the unique list. (ii) Non-singletons: all family members {F(P′)} except for P′ are erased from the list. (iii) Overlap to previously identified proteins: if P′ has one family-member Q that has already been included in the unique list at a previous step, the representative P′ and all other family members {F(P′)} except for Q will be removed from the stack. Note that this situation may have two reasons: (a) because of the asymmetrical nature of the distance-network generated by BLAST, and (b) due to some overlap between domains that invalidates the triangular relation (e.g. A similar to B and A similar to C does not imply that B is also similar to C). The algorithm completes if no protein remains in the stack.
User options
The user-defined parameter ‘smallest first’ or ‘largest first’ influences the final set of representatives in the following way: assume a set of three proteins with A and B being single domain non-homologous proteins and with C being a two-domain fusion of A+B. For a certain HSSP-value the setting ‘largest first’ would yield one group (A, B, C) whereas the setting ‘smallest first’ yields two (A,C) and (B,C). Sequence-space-hopping is a procedure to enlarge protein families by applying a triangular equation: if HV(A,B)>0, HV(B,C)>0 and HV(A,C)<0 this usually implies that we cannot infer the similarity between A and C directly (9,18). Sequence-space-hopping (or intermediate sequence searches) explore the fact that B is an intermediate common to families A and C to infer the similarity between A and C. We enable the user to apply this concept until no more new homologue sequences are found. ‘Smallest first’ often leads to families that can be connected via sequence-space-hopping. In our example an alignment of A would lead to sequence C and the second-round alignment of C would bring us back to A but also to B. Note: the default setting for the algorithm is ‘largest first’.
Output
Since our server accepts a range of HSSP-values instead of a single value in order to better exploit a once done BLAST-run on a submitted set, one output-file is produced for each HSSP-threshold processed by the program. Those output-files are simple FASTA-files each one of them holding a single representative set. When using the internet-version of UniqueProt, the output will be downloadable from our server in a compressed format (zip or tar) once the job has been finished. To get a better overview, user-friendly html-files with links to the mentioned FASTA-files can be obtained additionally and will be included in the compressed archive. These files will also contain the HSSP-values for each submitted protein-pair.
CONCLUSIONS
Although the program treats sequences as a whole rather than considering domains, the UniqueProt algorithm is a convenient and relatively fast way to thin out some set of sequences by removing bias originating from redundancy without losing the most important representatives. A data set containing ∼1000 sequences submitted to our server takes on average 15 min to complete. There is a restriction on the amount of data (500 kb for FASTA-files, 20 kb for ID-files, 10 Mb for alignment-files) in order to prevent overload of our CPU resources. Users who want to process larger sets can download the software and run it on their local Linux/Unix machines.
UniqueProt constitutes a level in between a relatively slow and careful clustering algorithm as used for example in GeneRAGE (19) and between the extremely fast and crude bias-reduction scheme CD-HI (20). We compared UniqueProt to the clustering method on a single data set of 187 nuclear-matrix associated proteins taken from SWISS-PROT. GeneRAGE grouped these proteins into 27 clusters. We grouped the same set through UniqueProt using different HSSP-values and both algorithm-modes (‘smallest first’ and ‘largest first’). We found the highest overlap between the two methods at an HSSP-value of 10 and with the mode ‘largest first’. Seventeen of 27 GeneRAGE clusters contained at least one representative in the mentioned UniqueProt set. The reason for the rather high value for the best-fit proximity threshold (HSSP-value of +10) was that GeneRAGE grouped half the proteins in the data set into one cluster and split the remaining proteins into many small clusters. Although, we have no good reason to assume that our single test is representative for all possible data sets, we were encouraged that UniqueProt is an alternative that works fast, is accessible and probably accurate enough if the proteins have similar domain architectures. We plan to investigate to what extent we could apply the fast algorithm employed in CD-HI (20) to achieve a first, fast grouping of our results in the future.
Acknowledgments
ACKNOWLEDGEMENTS
Thanks to Jinfeng Liu and Megan Restuccia (Columbia) for computer assistance and to Avner Schlessinger (Columbia) for testing the program over and over again. Thanks also to the anonymous reviewers for their help to improve manuscript and tool. This work was supported by the grants RO1-GM63029-01 from the National Institute of Health (NIH) and 1-R01-LM07329-01 from the National Library of Medicine (NLM). Last, but not least, thanks to Amos Bairoch (SIB, Geneva), Rolf Apweiler (EBI, Hinxton), Phil Bourne (San Diego University) and their crews for maintaining excellent databases and to all experimentalists who enabled this tool by making their data publicly available.
REFERENCES
- 1.Rost B. (2002) Enzyme function less conserved than anticipated. J. Mol. Biol., 318, 595–608. [DOI] [PubMed] [Google Scholar]
- 2.Todd A.E., Orengo,C.A. and Thornton,J.M. (2001) Evolution of function in protein superfamilies, from a structural perspective. J. Mol. Biol., 307, 1113–1143. [DOI] [PubMed] [Google Scholar]
- 3.Nair R. and Rost,B. (2002) Sequence conserved for sub-cellular localization. Protein Sci., 11, 2836–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamames J., Ouzounis,C., Casari,G., Sander,C. and Valencia,A. (1998) EUCLID: automatic classification of proteins in functional classes by their database annotations. Bioinformatics, 14, 542–543. [DOI] [PubMed] [Google Scholar]
- 5.Devos D. and Valencia,A. (2000) Practical limits of function prediction. Proteins, 41, 98–107. [PubMed] [Google Scholar]
- 6.Ashburner M., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S. and Eppig,J.T. (2000) Gene ontology: tool for the unification of biology. The gene ontology consortium. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson C.A., Kreychman,J. and Gerstein,M. (2000) Assessing annotation transfer for genomics: quantifying the relations between protein sequence, structure and function through traditional and probabilistic scores. J. Mol. Biol., 297, 233–249. [DOI] [PubMed] [Google Scholar]
- 8.Brenner S.E., Chothia,C. and Hubbard,T.J.P. (1998) Assessing sequence comparison methods with reliable structurally identified distant evolutionary relationships. Proc. Natl Acad. Sci. USA, 95, 6073–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rost B. (1999) Twilight zone of protein sequence alignments. Protein Eng., 12, 85–94. [DOI] [PubMed] [Google Scholar]
- 10.Yang A.S. and Honig,B. (2000) An integrated approach to the analysis and modeling of protein sequences and structures. II. On the relationship between sequence and structural similarity for proteins that are not obviously related in sequence. J. Mol. Biol., 301, 679–689. [DOI] [PubMed] [Google Scholar]
- 11.Alexandrov N.N. and Soloveyev,V.V. (1998) Statistical significance of ungapped sequence alignments. In Altman,R.B., Dunker,A.K., Hunter,L. and Klein,T.E. (eds), HICCS′ 98: Pacific Symposium on Biocomputing′ 98. World Scientific, Maui, Hawaii, USA, pp. 463–472. [PubMed] [Google Scholar]
- 12.Sander C. and Schneider,R. (1991) Database of homology-derived structures and the structural meaning of sequence alignment. Proteins, 9, 56–68. [DOI] [PubMed] [Google Scholar]
- 13.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berman H.M., Westbrook,J., Feng,Z., Gillliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul S.F. and Gish,W. (1996) Local alignment statistics. Methods Enzymol., 266, 460–480. [DOI] [PubMed] [Google Scholar]
- 16.Altschul S., Madden,T., Shaffer,A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D. (1997) Gapped Blast and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobohm U. and Sander,C. (1994) Enlarged representative set of protein structures. Protein Sci., 3, 522–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park J., Teichmann,S.A., Hubbard,T. and Chothia,C. (1997) Intermediate sequences increase the detection of distant sequence homologies. J. Mol. Biol., 273, 349–354. [DOI] [PubMed] [Google Scholar]
- 19.Enright A.J. and Ouzounis,C.A. (2000) GeneRAGE: a robust algorithm for sequence clustering and domain detection. Bioinformatics, 16, 451–457. [DOI] [PubMed] [Google Scholar]
- 20.Li W., Jaroszewski,L. and Godzik,A. (2001) Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics, 17, 282–283. [DOI] [PubMed] [Google Scholar]