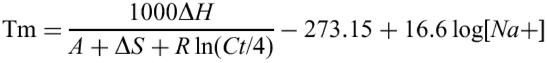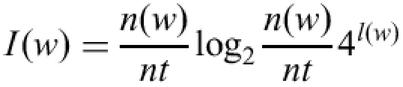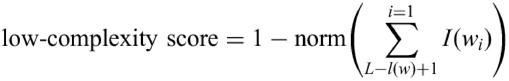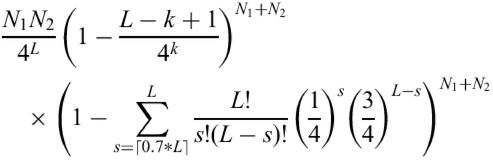Abstract
Optimal design of oligonucleotides for microarrays involves tedious and laborious work evaluating potential oligonucleotides relative to a series of parameters. The currently available tools for this purpose are limited in their flexibility and do not present the oligonucleotide designer with an overview of these parameters. We present here a flexible tool named OligoWiz for designing oligonucleotides for multiple purposes. OligoWiz presents a set of parameter scores in a graphical interface to facilitate an overview for the user. Additional custom parameter scores can easily be added to the program to extend the default parameters: homology, ΔTm, low-complexity, position and GATC-only. Furthermore we present an analysis of the limitations in designing oligonucleotide sets that can detect transcripts from multiple organisms. OligoWiz is available at www.cbs.dtu.dk/services/OligoWiz/.
INTRODUCTION
Oligonucleotides of 50–70 bp, or even shorter in the range of 20–30 bp, are now widely used for microarrays in gene expression studies. An ideal oligonucleotide must discriminate well between its intended target and all other targets in the pool. It must detect concentration differences under the hybridization conditions with minimal variation and detect within the range of its target concentration under the given conditions. However in reality the optimal oligonucleotide will often have to be a compromise between these criteria.
Here we present a flexible program for designing optimal oligonucleotides for microarrays named OligoWiz. The program will allow for the design of highly specialized arrays as well as general use arrays. OligoWiz evaluates and graphically presents all potential oligonucleotides in the input sequences according to a collection of parameters, each of which can be assigned a different weight. In addition to a standard collection of parameters (homology, ΔTm, GATC-only, position within transcript and complexity), custom parameters may be of relevance to specialized microarrays. Therefore OligoWiz has been designed to be flexible towards additional user defined parameters. We present an analysis of the limitations in designing oligonucleotide sets that can detect transcripts from multiple organisms.
OligoWiz is implemented as a client–server solution. The server is responsible for the calculation of the scores. The client is used for submitting jobs to the server as well as for visualizing the scores and fine-tuning the placement of the oligonucleotides. The OligoWiz client is freely available from the OligoWiz web page: http://www.cbs.dtu.dk/services/OligoWiz/ and the server is commercially available.
METHOD
The OligoWiz client is written in Java 1.3.1 with cross-platform functionality in mind. It is tested on MacOS X, Linux and Windows, and should be able to run on any platform with Java 1.3.1 or better.
The OligoWiz server was developed on a SGI Unix system and is written in Perl5. The server utilizes the BLAST program for homology search (1) and saco_patterns for generating a pattern information database (2). The server program is parallelized using the Perl module ChildManager. Additional information about the sequence databases available for OligoWiz can be found on the OligoWiz web page.
Multi-transcriptome analysis
Predicted genes from two strains of Campylobacter jejuni (RM1221 from TIGR and NCTC11168 from GenBank) were extracted based on EasyGene annotations with R-values <2 (Larsen and Krogh, manuscript submitted). The rat, mouse, Streptococcaceae pneumoniae and Streptococcaceae pyogenes coding DNA sequences (CDS) were extracted based on the ftp://ftp.ncbi.nih.gov/genbank/genomes/M_musculus/RNA/rna.gbk.gz, ftp://ftp.ncbi.nih.gov/genbank/genomes/R_norvegicus/rn_mrna.gz files, NC_003028 and NC_002737 annotations, respectively.
Patterns were extracted and redundancy reduced for stretches of 15 bp using saco_patterns (2) and custom Perl scripts. The oligonucleotides were further redundancy reduced using BLAST (1). The oligonucleotides were mapped back on the transcriptomes using Perl scripts.
DESIGNING OLIGONUCLEOTIDES
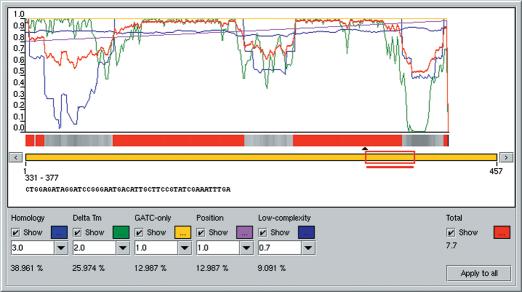
The protocol for oligonucleotide design using OligoWiz is as follows: the program calculates a series of independent parameter scores that describes all potential oligonucleotides. Each parameter score has a value between 0 and 1, where 1 is optimal. For each oligonucleotide, the parameter scores are weighted and summed to form a total score, which is normalized to a value between 0 and 1. The graphical interface presents the user with graphs of all the parameter scores including the total score, along a virtual sequence (Fig. 1). OligoWiz points out the oligonucleotide with the highest total score in each input sequence, but the graphical interface also allows for direct user intervention. In addition to the scores the user interface can also visualize sequence annotations in a color bar. An example could be exon/intron annotation. The user can then select an oligonucleotide based on the information presented. Below we describe the parameter scores that are used for the oligonucleotide design.
Figure 1.
Screenshot of part of OligoWiz's graphical interface. The graphs demonstrate the score of oligonucleotides along the length of the transcript sequence. The red graph (total) represents the total weighted score from which the automated oligonucleotide selection is based. The red box placed on the orange bar is a handle for manual oligonucleotide selection. Below the graphs the weights for the parameter scores can be set. Shown is the S.cerevisiae transcript AA417464.
Detailed descriptions of how to use the program are found at: www.cbs.dtu.dk/services/OligoWiz/instructions.html.
Cross-hybridization
In order for the oligonucleotide to avoid cross-hybridization, the affinity difference between the intended target and all other targets should ideally be maximized. However, such a calculation requires that the concentration of all targets are known, which they rarely are. Even if the concentrations were ignored, an affinity calculation would be very time consuming for large collections of oligonucleotides. In practice, a homology search between all targets and the oligonucleotide is a good estimate of the affinity of the oligonucleotide to alternative targets. Experimental evidence suggests that a significantly false signal can be detected if a 50 bp oligonucleotide has >75–80% of the bases complementary or if continuous stretches of >15 bp are complementary to a false target (3). Similar results have been found for 60mers (4).
To estimate the degree of cross-hybridization, OligoWiz calculates a homology score for each oligonucleotide in a given sequence, based on a BLAST search of the input sequence against a species-specific database. The resulting BLAST hits are then evaluated relative to each oligo along the input sequence.
Let m be the number of BLAST hits considered in position i of the oligonucleotide and h={h1i,…, hmi} be the BLAST hits in position i.
where L is the length of the oligonucleotide. Oligonucleotides with 100% identity to any considered BLAST hit along the full length of the oligonucleotide will get a score of 0, in contrast to the score value 1 assigned to oligonucleotides that have no homology to any considered BLAST hit. Homology is, in this context, used to define sequence identity and not necessarily evolutionary relationship.
To avoid including BLAST hits against the gene sequence that the oligonucleotide originated from OligoWiz allows for filtering against BLAST hits that cover more than a fraction (default 0.8) of the entire length of the input sequence and have a homology higher than a threshold (default 97%). The filter also enables filtering of homology lower than a threshold (default 70%) or shorter than a given length (default 15 bp).
ΔTm
To further enhance the ability of the oligonucleotides to discriminate between the targets, the hybridization and washing conditions need to be optimal. Therefore it is crucial that all oligonucleotides perform well under similar hybridization conditions. The melting temperature of the DNA:DNA duplex (Tm) is a good description of an oligonucleotide hybridization property and as such the minimal difference between the Tm of the oligonucleotides is desired.
OligoWiz uses a nearest-neighbor model for Tm estimation:
where ΔH is the enthalpy, ΔS is the entropy change of the nucleation reaction; A is a constant correcting for helix initiation (−10.8), R is the universal gas constant (1.99 cal K−1 mol−1), Ct is the total molar concentration of strands (5). Since the total molar concentration of strands is unknown for most microarray experiments, OligoWiz uses a constant of 2.5×10−10 M. Based on the Tm estimation a ΔTm score is calculated.
where OTm by default is the mean Tm of all oligonucleotides in all input sequences of aim length (user specified) or a specific user specified optimal Tm.
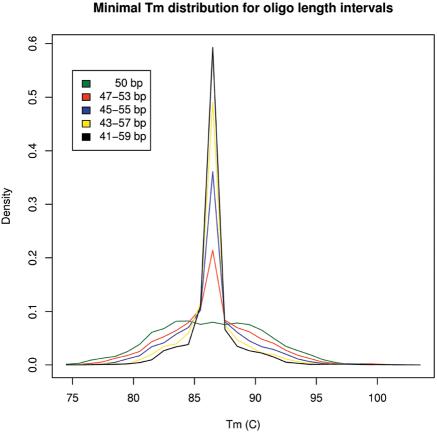
To facilitate maximal freedom in selecting oligonucleotides according to other requirements, OligoWiz calculates melting temperatures for all oligonucleotides in a user defined length interval. For each 5′ position along the input sequence the oligonucleotide length (extending toward the 3′ end) with the best ΔTm score is chosen. This narrows the Tm difference distribution of the oligonucleotides significantly as can be seen in Figure 2. Thus, the ΔTm score and oligonucleotide length are connected. The oligonucleotide length influences the remaining score calculations (except the position score). Therefore the ΔTm score is the first calculation the OligoWiz server performs.
Figure 2.
The Tm distribution for oligonucleotides with minimal Tm difference within different length intervals. For each position in 6600 S.cerevisiae transcripts, the oligonucleotide with the smallest Tm difference within the given length interval was selected. By selecting oligonucleotides in this way OligoWiz optimizes the number of oligonucleotides with a small Tm difference.
Position within transcript
The position within the target transcript can be of importance, especially for long transcripts. The reverse transcriptase will fall off the transcript with a certain probability and the further away from the starting point the less signal will be generated. Therefore oligonucleotides with target positions closer to the starting point of the reverse transcriptase are preferable.
If the labeling commences from the 3′ end (poly A tail) the following score is used:
where dp is the probability that the reverse transcriptase will fall off its template at any given base and Δ3′end is the oligonucleotide distance to the 3′ end of the input sequence.
In cases where the labeling is done with random primers, as would be the case under prokaryote mRNA labeling, the chance of having an oligonucleotide upstream of a given position should be accounted for:
where c is a constant indicating the probability that a random primer will bind at any given position.
Low-complexity filtering
To avoid oligonucleotides composed of very common sequence fragments in probe design a low-complexity score was implemented. Different sequences are common in different species, therefore to estimate a low-complexity measure for an oligonucleotide a list of sequence subfragments and the information content of these is generated specifically for each species. The information content can be calculated by the following equation (6):
where n(w) is the number of occurrences of a pattern in the transcriptome, l(w) the pattern length, nt is the total number of patterns found of a given length. OligoWiz uses this list to calculate a low-complexity score for each oligonucleotide:
where L is the length of the oligonucleotide, wi is the pattern in position i and norm is a function that normalizes the summed information to a value between 1 and 0.
A low-complexity score of 0 indicates an oligonucleotide with very low complexity. By far the majority of oligonucleotides have a low-complexity score between 1 and 0.8, and in fact the distribution is exponential. As a consequence the low-complexity score only significantly influences few oligonucleotides, with very low low-complexity measures.
GATC-only score
To allow for filtering out sequence containing ambiguity annotation OligoWiz has a score called ‘GATC-only’. Oligonucleotides containing R, Y, M, K, X, S, W, H, B, V, D, N or anything else but A, C, T or G will be given a score of 0, while oligonucleotides only containing G, A, T and C will be assigned a score of 1.
Custom parameter scores
In addition to the parameter scores calculated by the OligoWiz server, custom scores can be added to the list and will be handled by the graphically interface. (For detailed description see the program documentation.)
RESULTS
To evaluate OligoWiz, oligonucleotides were designed to detect the expression from the 6600 genes annotated in the Saccharomyces cerevisiae genome (7). The design was performed using standard settings of OligoWiz, designing oligonucleotides in the length interval 45–55 bp. The homology search and complexity score was based on whole genome databases. The mean Tm of the oligonucleotides was 75.7°C. OligoWiz was able to find oligonucleotides with good parameters for almost all genes as shown in Table 1.
Table 1. Properties of oligonucleotides for 6600 S.cerevisiae transcripts designed by OligoWiz.
| Homology | ΔTm | Position |
|---|---|---|
| 6112 No homology | 6373 <±1°C | 4345 <100 bp from 3′ end |
| 161 <70% | 67 <±2°C>±1°C | 1800 100–200 bp |
| 47 70–80% | 22 <±3°C>±2°C | 344 200–300 bp |
| 52 80–90% | 13 <±5°C>±3°C | 57 300–400 bp |
| 232 >90% | 113 >±5°C | 42 >400 bp |
In the ‘Homology’ column the number of oligonucleotides with the given degree of homology is listed. In the ‘ΔTm’ column the number of oligonucleotides within the given Tm range from the mean. In the ‘Position’ column the number of oligonucleotides within the given distance from the 3′ end of the transcript. The oligonucleotides were designed using default settings and S.cerevisiae databases.
Less than 150 oligonucleotides had low complexity. The calculations of the parameters for the 6600 yeast genes were done in just 20 min.
Multi-transcriptome arrays
Microarray experiments using oligonucleotides covering a whole transcriptome are still very expensive to initiate and it is of course best to reduce these costs if possible. One possible way to reduce the setup cost for several organisms is to design oligonucleotides that enable detection of transcripts from more than one organism. Furthermore a microarray that has been designed to be robust toward strain differences is of interest for laboratories that focus on more than one strain of a species. To shed light on this topic the following analysis was engaged.
If it is assumed that the sequences in two transcriptomes are random and independent, the expected number of oligonucleotides of length L that will be perfectly complementary to a sequence in each of two transcriptomes of lengths N1 and N2 would be approximately N1 * N2/4L. If we further restrict the oligonucleotides not to have more than 70% of the sequence in common with any other sequence within the transcriptomes and to be unique in any stretch of length k (k being less than 70% of L). Then the expected number of oligonucleotides that meet these criteria can be approximated by:
Here we neglect the chance of an oligonucleotide sharing k continuous bases with more than one other transcript and assume that the transcript wherein we find an oligonucleotide is very short relative to the total transcriptome.
A shared 50mer where any of its 36 possible 15mers are unique within each transcriptome and overall does not share >70% homology to any other 50mer (3), is unlikely even in large transcriptomes like the mouse and rat transcriptomes. Furthermore, we find that the expected number of such shared oligonucleotides in random transcriptomes is very dependent on the length of the oligonucleotide.
However, transcriptomes are not composed of random sequences and related transcriptomes are not independent. To investigate the impact of such biases the number of 50mers in the transcriptomes of rat and mouse as well as in S.pneumoniae and S.pyogenes and the two C.jejuni strains (NCTC11168 and RM1221) were counted. Hereof only few were shared within the Streptococcaceae (<0.1%) and the rodents (<2%) species relative to the 55–59% between the two C.jejuni strains, indicating how related the two C.jejuni strains are. Still all the compared groups shared far more oligonucleotides than expected for random transcriptomes with the respective sizes.
The theoretical model predicts that only a very moderate fraction of the oligonucleotides will be rejected by the redundancy criteria. In reality we find that 73, 34 and 11% of the Streptococcaceae, C.jejuni and rodent oligonucleotides pass the redundancy criteria respectively (i.e. the oligonucleotide does not share any stretch with continuous complementary of 15 bp or more, and have <70% overall complementary, to any other transcript sequence from either transcriptome). This is considerably less than expected for a random sequence, indicating some redundancy in the sequences. The differences in redundancy between the groups may to some degree be explained by the AT skew in the C.jejuni transcriptomes (68%) and the large size of the rodent transcriptomes.
The shared and non-redundant 50mers were mapped back on the transcriptome and the transcripts that could be measured by these were counted. A total of 619 out of the 9538 and 9487 transcripts in mouse and rat, respectively, and only 23 transcripts out of the 2094 and 1697 S.pneumoniae and S.pyogenes transcriptomes, respectively. In the two highly related C.jejuni strains on the other hand 88 and 76% or 1437 transcripts out of 1640 and 1880 were found to be measurable by common oligonucleotides.
To investigate the potential for shorter oligonucleotides, all oligonucleotides of 25 bp in the transcriptome pairs were likewise counted. As expected more conserved 25mers than 50mers were found, especially between the more distantly related Streptococcaceae species (Table 2). Using the same redundancy criteria as for the 50mers reduced the potential numbers of 25mers relatively less than was the case for the 50mers. This is expected as a 25mer consists of only 11 stretches of 15 bp, in contrast to the 36 stretches in a 50mer. The shared and non-redundant 25mers were mapped back on the transcriptome and the transcripts that could be measured by these were counted (Table 3). Approximately twice as many rodent and more than seven times more Streptococcaceae transcripts were measurable by common 25mers.
Table 2. The total number of oligonucleotides of 50, 25 and 18 bp length and the number of annotated transcript in the respective organisms.
| Organism | Total 50mers | 25mers | 18mers | Transcripts |
|---|---|---|---|---|
| Rat | 11 089 538 | 11 001 241 | 10 902 992 | 9487 |
| Mouse | 12 712 223 | 12 563 540 | 12 408 777 | 9538 |
| S.pneumoniae | 1 635 198 | 1 679 397 | 2094 | |
| S.pyogenes | 1 455 453 | 1 493 553 | 1697 | |
| C.jejuni NCTC11168 | 1 432 822 | 1 481 672 | 1640 | |
| C.jejuni RM1221 | 1 533 292 | 1 594 699 | 1880 |
Table 3. Number of oligonucleotides found within the three groups.
| Group | Shared 50mers | Non-redundant 50mers | Transcripts measurable by common 50mers | Shared 25mers | Non-redundant 25mers | Transcripts measurable by common 25mers |
|---|---|---|---|---|---|---|
| Rodent | 200 333 | 23 017 | 619 | 363 105 | 183 255 | 1238 |
| Streptococcaceae | 1321 | 967 | 23 | 5427 | 4638 | 174 |
| C.jejuni | 848 905 | 290 283 | 1493 | 1 058 108 | 696 842 | 1546 |
Finally, we investigated the potential of even shorter oligonucleotides of 18 bp to allow for designing multi-transcriptome arrays for mouse and rat. It was possible to detect 2901 transcripts with common 18mers.
The shorter oligonucleotides enabled more transcripts to be measured by common oligonucleotides, but the impact of the oligonucleotide length does not have the impact expected based on the random transcriptome model. This suggests that the majority of the common oligonucleotides originate from long stretches that are conserved between the transcriptomes.
To demonstrate the flexibility of OligoWiz, we added a custom score indicating oligonucleotides that were suitable for multi-transcriptome arrays. The custom score was assigned a 1 where oligonucleotides were shared and not redundant between two transcriptomes, otherwise a 0 was assigned. The custom scores were added as an extra column in the input file for the OligoWiz client program (ASCII file) and the score was named in the file header. The OligoWiz client automatically included the new parameter and allowed for oligonucleotide selection using the new parameter.
DISCUSSION
The design of oligonucleotides for microarray analysis is a laborious and tedious work if the analysis is not automated in some way. A couple of tools for this purpose already exist (8–10). Li and Stormo (8) have developed a program that focuses on avoiding cross-hybridization. OligoArray by Rouillard et al. (9) is, as OligoWiz, a client server application that runs locally and sends jobs to a web server. However OligoArray requires a local BLAST function. Mrowka et al. (10) describes a method to design oligonucleotides for the human ENSEMBL project entries. Neither of these tools supports extensive user interaction and they give only limited overview of the design process.
The purposes of microarrays are numerous and, in many cases, this has an impact on the design. OligoWiz is designed to have maximum degrees of freedom and to give the designer an overview of a number of parameters in a graphical display. In particular, OligoWiz gives the designer extended freedom by minimizing the ΔTm of the oligonucleotides by varying their length within an interval.
Additional custom parameters can easily be added to the parameter collection and displayed in the graphical display. This may be a phred score to indicate sequence quality (11); indications of intron or exon regions or some other parameter the designer finds important for the microarray design at hand.
The amount of shared oligonucleotides between the three transcriptome pairs (mouse/rat, S.pneumoniae/S.pyogenes and two C.jejuni strains NCTC11168/RM1221) indicates how related the transcriptomes are (Table 2). This by far exceeds the amount expected for random transcriptomes. However, the fraction of oligonucleotides shared between the transcriptomes is a major bottleneck for the success of multi-transcriptome array design. Only relatively few potential multi-transcriptome oligonucleotides are rejected because of redundancy: 27–66% in the two bacterial groups and 89% in the larger rodent transcriptomes. For transcriptomes with many paralogs this may be of greater importance. To the extent this is caused by paralogs, grouping paralogs, and thereby accepting not to be able to distinguish these in the resulting array, can diminish loss of potential oligonucleotide fragments.
Our analysis also indicates the impact the length of the oligonucleotides has on the possibility for designing multi-transcriptome microarrays. Furthermore, shorter oligonucleotides may be better at discriminating between perfect complementary sequence and sequences with mismatches. This is indicated by the increasing difference in melting temperature between such sequences, as they get shorter (data not shown).
Alternative hybridization chemistries that will allow for shorter oligonucleotides to be used as microarray probes like LNA (reviewed in 12) may allow for multi-transcriptome arrays of more distantly related organisms. However our analysis indicates the strong limitations this concept has. But, for very related organisms, it is possible to save a number of oligonucleotides and make the array measurements more strain robust for a considerable number of genes.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank L. Jensen, A. Fausbøll, U. de Lichtenberg and T. Jensen for discussion on the theoretical multi-transcriptome model; Peter Hallin for discussions on multi-transcriptome design, and David Ussery for proofreading the manuscript. This work was funded by grants from Novozymes A/S, The Danish National Research Foundation and The Danish Biotechnology Instrument Center.
REFERENCES
- 1.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 2.Jensen L.J. and Knudsen,S. (2000) Automatic discovery of regulatory patterns in promoter regions based on whole cell expression data and functional annotation. Bioinformatics, 16, 326–333. [DOI] [PubMed] [Google Scholar]
- 3.Kane M.D., Jatkoe,T.A., Stumpf,C.R., Lu,J., Thomas,J.D. and Madore,S.J. (2000) Assessment of the sensitivity and specificity of oligonucleotide (50mer) microarrays. Nucleic Acids Res., 28, 4552–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes T.R., Mao,M., Jones.A.R., Burchard,J., Marton,M.J., Shannon,K.W., Lefkowitz,S.M., Ziman,M., Schelter,J.M., Meyer,M.R. et al. (2001) Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol., 19, 342–347. [DOI] [PubMed] [Google Scholar]
- 5.Breslauer K.J., Frank,R., Blocker,H. and Marky,L.A. (1986) Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA, 83, 3746–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shannon C.E. (1948) The mathematical theory of communication. Bell System Tech. J., 27, 379–423. [Google Scholar]
- 7.Goffeau A., Barrell,B.G., Bussey,H., Davis,R.W., Dujon,B., Feldmann,H., Galibert,F., Hoheisel,J.D., Jacq,C., Johnston,M. et al. (1996) Life with 6000 genes. Science, 274, 563–567. [DOI] [PubMed] [Google Scholar]
- 8.Li F. and Stormo,G.D. (2001) Selection of optimal DNA oligos for gene expression arrays. Bioinformatics, 17, 1067–1076. [DOI] [PubMed] [Google Scholar]
- 9.Rouillard J.M., Herbert,C.J. and Zuker,M. (2002) OligoArray: genome-scale oligonucleotide design for microarrays. Bioinformatics, 18, 486–487. [DOI] [PubMed] [Google Scholar]
- 10.Mrowka R., Schuchhardt,J. and Gille,C. (2002) Oligodb-interactive design of oligo DNA for transcription profiling of human genes. Bioinformatics, 18, 1686–1687. [DOI] [PubMed] [Google Scholar]
- 11.Ewing B. and Green,P. (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res., 8, 186–194. [PubMed] [Google Scholar]
- 12.Braasch D.A. and Corey,D.R. (2001) Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem. Biol., 8, 1–7. [DOI] [PubMed] [Google Scholar]