Abstract
MAVID is a multiple alignment program suitable for many large genomic regions. The MAVID web server allows biomedical researchers to quickly obtain multiple alignments for genomic sequences and to subsequently analyse the alignments for conserved regions. MAVID has been successfully used for the alignment of closely related species such as primates and also for the alignment of more distant organisms such as human and fugu. The server is fast, capable of aligning hundreds of kilobases in less than a minute. The multiple alignment is used to build a phylogenetic tree for the sequences, which is subsequently used as a basis for identifying conserved regions in the alignment. The server can be accessed at http://baboon.math.berkeley.edu/mavid/.
INTRODUCTION
The comparison of the mouse and human genomes (1) has demonstrated the power of comparative genomics in inferring the evolutionary history of species and in identifying functional regions in genomes. The possibilities for identifying regions under selection are enhanced with the addition of more sequences and this observation has led to numerous ‘focused sequencing’ projects which seek to obtain sequence for a small region of a genome in numerous other organisms (2).
Biologists who seek to analyse conserved regions among homologous sequences are faced with the daunting task of aligning large genomic regions and subsequently sifting through massive amounts of data. In order to facilitate the discovery process without requiring biologists to download and install complex software, a number of web servers for alignment and analysis have been set up in recent years (3,4). These servers align submitted sequences and then generate plots or graphs designed to help researchers identify conserved regions. A major drawback of existing servers which support the alignment of large genomic sequences is that they can only perform pairwise sequence comparisons (some servers allow for the input of multiple sequences, but only perform pairwise comparisons).
We have developed a web server to allow researchers to obtain multiple alignments for homologous sequences from multiple genomes and to extract meaningful information from the alignments. The web server uses the MAVID alignment program, which is able to quickly and accurately align large genomic regions. The program is also efficient in processing large numbers of sequences and is therefore also suitable for the alignment of mitochondrial sequences, viral genomes and other data sets for which there are many sequences.
We have designed the server with biomedical researchers in mind. At times, flexibility has been sacrificed for clarity and transparency, but the advantage is that different sequences can be easily aligned without the need to set parameters and otherwise interact with the server. For example, human, mouse and rat BACs can be aligned just as easily as chicken and fugu sequences, or hundreds of HIV sequences. The output is organized in such a way that conserved regions between subsets of sequences can be quickly identified for further investigation. Multiple alignments are provided in a variety of convenient formats which facilitate visual checking of the alignments and at the same time enable more sophisticated checking of consistency and accuracy.
The web server is freely accessible and privacy of users is ensured by not requesting user email addresses or other information.
METHOD
MAVID is a progressive alignment program (Bray and Pachter, manuscript submitted). The program works by recursively aligning the ‘alignments’ at ancestral nodes of the guide tree. At each internal node, ancestral sequences are inferred from the existing alignments using maximum likelihood and these alignments are then aligned using the AVID program (5).
The server goes through a number of steps:
Sequences are repeat-masked using the DUST program (Tatusov and Lipman, unpublished).
A random (almost complete) binary guide tree is generated for alignment of the sequences using the progressive alignment method.
The sequences are aligned using MAVID.
A phylogenetic tree is inferred from the multiple alignment using the neighbor joining method.
Steps 3 and 4 are repeated for a total of three iterations.
Pairwise alignments are generated from the multiple alignment with respect to all of the sequences and these are used to generate conservation plots and to identifiy conserved regions.
FEATURES
The MAVID server supports a number of functions that are useful for biomedical researchers. Input to the server consists solely of a set of sequences in multi-FASTA format; prior knowledge of the phylogenetic tree relating the sequences is not required. Output (Fig. 1) includes the multiple alignment, phylogenetic tree and ATV applet for visualization (6), visualization of the induced pairwise alignments in VISTA format (3) and conserved (not just similar) regions. Results are saved on a web page which users can bookmark for future reference; sequences can also be submitted anonymously.
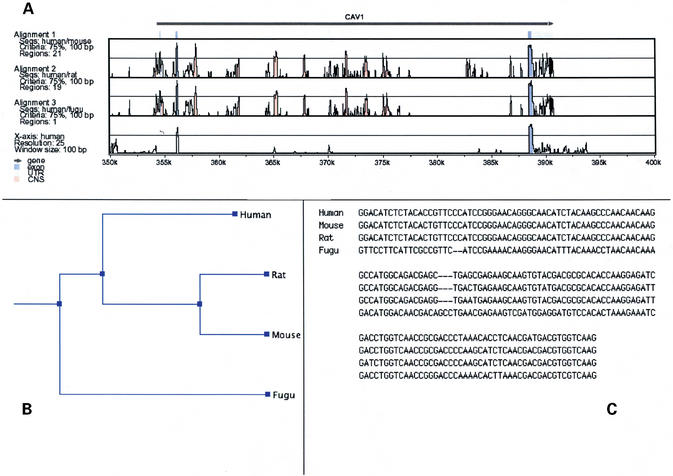
Figure 1.
Examples of output from the MAVID server for four sequences (human, mouse, rat and fugu) containing the CAV1 gene. (A) Pairwise VISTA plots from the projection of the multiple alignment onto the human sequence. (B) The phylogenetic tree constructed from the alignment. (C) A region from the multiple alignment.
REFERENCES
- 1.Waterston R.H., Lindblad-Toh,K., Birney,E., Rogers,J., Abril,J.F., Agarwal,P., Agarwala,R., Ainscough,R., Alexandersson,M., An,P. et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature, 420, 520–562. [DOI] [PubMed] [Google Scholar]
- 2.Boffelli D., McAuliffe,J., Ovcharenko,D., Lewis,K., Ovcharenko,I., Pachter,L. and Rubin,E.M. (2003) Phylogenetic shadowing of primate sequences to find functional regions in the human genome. Science, 299, 1391–1394. [DOI] [PubMed] [Google Scholar]
- 3.Mayor C., Brudno,M., Schwartz,J.R., Poliakov,A., Rubin,E.M., Frazer,K.A., Pachter,L. and Dubchak,I. (2000) VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics, 16, 1046–1047. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz S., Zhang,Z., Frazer,K.A., Smit,A., Riemer,C., Bouck,J., Gibbs,R., Hardison,R. and Miller,W. (2000) PiPMaker—a web server for aligning two genomic DNA sequences. Genome Res., 10, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bray N., Dubchak,I. and Pachter,L. (2003) AVID: a global alignment program. Genome Res., 13, 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zmasek C.M. and Eddy,S.R. (2001) ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics, 17, 383–384. [DOI] [PubMed] [Google Scholar]