Abstract
Onto-Tools is a set of four seamlessly integrated databases: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Onto-Express is able to automatically translate lists of genes found to be differentially regulated in a given condition into functional profiles characterizing the impact of the condition studied upon various biological processes and pathways. OE constructs functional profiles (using Gene Ontology terms) for the following categories: biochemical function, biological process, cellular role, cellular component, molecular function and chromosome location. Statistical significance values are calculated for each category. Once the initial exploratory analysis identified a number of relevant biological processes, specific mechanisms of interactions can be hypothesized for the conditions studied. Currently, many commercial arrays are available for the investigation of specific mechanisms. Each such array is characterized by a biological bias determined by the extent to which the genes present on the array represent specific pathways. Onto-Compare is a tool that allows efficient comparisons of any sets of commercial or custom arrays. Using Onto-Compare, a researcher can determine quickly which array, or set of arrays, covers best the hypotheses studied. In many situations, no commercial arrays are available for specific biological mechanisms. Onto-Design is a tool that allows the user to select genes that represent given functional categories. Onto-Translate allows the user to translate easily lists of accession numbers, UniGene clusters and Affymetrix probes into one another. All tools above are seamlessly integrated. The Onto-Tools are available online at http://vortex.cs.wayne.edu/Projects.html.
INTRODUCTION
Microarrays are at the center of a revolution in biotechnology, allowing researchers to screen tens of thousands of genes simultaneously, generating a staggering amount of data. The current challenge is to analyze these data and translate them into an understanding of the underlying biological phenomenon. A microarray experiment can be broadly divided into two steps. The first step is usually an exploratory search in which one tries to identify a subset of genes that may be playing an important role and formulate a hypothesis about the phenomenon studied. The second step usually is a very focused research that usually involves a small number of pathways and processes as required by the hypothesis proposed. Typically, the result of the first exploratory step is a set of differentially regulated genes. A major challenge is to translate this set of differentially regulated genes into a better biological understanding of the phenomenon that would allow a subsequent formulation of research hypotheses. This is usually accomplished by a tedious search of the literature and various online genomic databases such as NCBI, EMBL and DDBJ. Searching various online databases is an enormous task as different databases refer to the same piece of information differently and complementary information about the same gene may be stored in many different databases.
After finding a set of differentially regulated genes and formulating various hypotheses based on such genes, the research usually focuses on a small number of biological processes believed to be highly relevant. However, in many cases, even a small number of biological processes from few pathways may still involve hundreds of genes thus making microarrays the preferred tool. This focused research is best carried out by using an appropriately focused microarray that contains a set of genes that are only related to the problem at hand. Literally tens of focused commercial microarrays are available today. Some pathways are covered by several competing commercial microarrays using different sets of genes. Each such microarray will exhibit a biological bias determined by the choice of the particular genes present on the array. Furthermore, in spite of the large number of custom arrays currently available, not every possible biological problem will have a commercial array available. In many cases, a researcher may need, or choose, to design a microarray that is appropriate for testing their hypothesis.
This paper describes the Onto-Tools annotation databases together with a set of ontology-based tools that help address the problems identified above. Onto-Express (OE) is a tool designed to mine the available functional annotation data and help the researcher find relevant biological processes. Many months of tedious and inexact manual searches are substituted by a few minutes of fully automated analysis. Onto-Compare (OC) helps researchers analyze the biological bias of various commercial microarrays in order to find the array, or combination of arrays, that is best suited to investigate a given biological hypothesis. If one cannot find a suitable commercial microarray, Onto-Design (OD) is a tool that allows to quickly design a microarray by constructing an optimal set of genes for a given set of biological processes or pathways. Finally, Onto-Translate is a utility that allows quick conversions among a list of probe identifiers (IDs), accession numbers or cluster IDs. These tools are freely available at: http://vortex.cs.wayne.edu/Projects.html.
MATERIALS AND METHODS
Onto-Express
Microarrays have been introduced as powerful tools able to screen a large number of genes in an efficient manner. The typical result of a microarray experiment is a number of gene expression profiles, which in turn are used to generate hypotheses and locate effects on many, perhaps unrelated pathways. This is a typical hypothesis generating experiment. For this purpose, it is best to use comprehensive microarrays, that represent as many genes of an organism as possible. Currently, such arrays include tens of thousands of genes. For example, the HGU133 (A+B) set from Affymetrix Inc. contains 44 928 probes that represent 42 676 unique sequences from GenBank database corresponding to 28 036 UniGene clusters.
Typically, after conducting a microarray experiment, independent of the platform and the analysis methods used, one selects a set of genes that are found to be differentially expressed. These lists of differentially regulated genes need to be translated into biological processes or molecular functions characterizing the underlying biological phenomenon. This poses a requirement to analyze the genes from a functional point of view. Typically, in order to analyze a set of genes and create their functional profiles, one needs to search the literature and the various online databases. For example, a typical analysis of a set of differentially regulated genes will involve searching NCBI UniGene (1,2) and LocusLink (3) databases for each of the genes in the list. This is an extremely tedious and error-prone process. Furthermore, carrying out these manual searches in a systematic manner and finding out a simple frequency of a given biological process among the differentially regulated genes may produce misleading results (4).
Onto-Express (OE) (4,5) is one of the annotation databases integrated in Onto-Tools. OE is a tool designed to mine the available functional annotation data and help the researcher find relevant biological processes (4,5). Many months of tedious and inexact manual searches are substituted by a few minutes of fully automated analysis. The result of this analysis is a functional profile of the condition studied. In the latest version, this functional profile is accompanied by the computation of significance values for each functional category. Such values allow the user to distinguish between significant biological processes and random events. OE's utility has been demonstrated by analyzing data from a recent breast cancer study.
The input to OE is a list of GenBank accession numbers, Affymetrix probe IDs or UniGene cluster IDs. A functional category can be assigned to a gene based on specific experimental evidence or by theoretical inference (e.g. similarity with a protein having a known function). OE shows explicitly how many genes in a category are supported by experimental evidence (labelled ‘experimented’) and how many are inferred (‘inferred’). Those genes for which this information is not available are labelled ‘non-recorded’. The results are provided in graphical form and emailed to the user on request. OE constructs a functional profile for each of the Gene Ontology (GO) categories: cellular component, biological process and molecular function as well as biochemical function and cellular role, as defined by Proteome (http://www.incyte.com/sequence/proteome). As biological processes can be regulated within a local chromosomal region (e.g. imprinting), an additional profile is constructed for the chromosome location.
The probability model best suited to calculate the significance values would use a hypergeometric distribution (4). For a typical microarray experiment when the number of genes on the chip N≃10 000 and the number of selected genes is K≃100=1%N, the binomial approximates well the hypergeometric and, therefore, the hypergeometric was not implemented. The χ2 was also proposed for similar problems (6). Finally, Fisher's exact test is required when the sample size is small and the chi-square test cannot be used. OE provides implementations of the χ2 test, Fisher's exact test as well as the binomial test. The user can select between the binomial and the χ2 test. If χ2 is chosen, the program automatically calculates the expected values and uses Fisher's exact test when χ2 becomes unreliable (expected values <5).
Onto-Compare
Many microarray users embark upon ‘hypotheses generating experiments’ in which the goal is to find subsets of genes differentially regulated in a given condition. However, another major application of this type of data mining is in experiment design. An alternative to the ‘hypotheses generating experiments’ is the ‘hypothesis driven experiments’ in which one first constructs a hypothesis about the phenomenon under study and then performs directed experiments to test the hypothesis. However, specific hypotheses and a small number of pathways may still involve hundreds of genes. This is still too many for RT–PCRs, western blotting and other gene specific techniques, so the microarray technology is still the preferred approach.
Currently, no two arrays offer exactly the same set of genes. When a hypothesis of a certain mechanism does exist, we argue that one should use the array(s) that best represent the corresponding pathways. This can be accomplished by analyzing the list of genes on all existing arrays and providing information about the pathways and biological mechanisms covered by the genes on each array. If array A contains 10 000 genes but only 80 are related to a given pathway and array B contains only 400 genes but 200 of them are related to the pathway of interest, the experiment may provide more information if performed with array B instead of A. This can also translate into significant cost savings.
Many commercial microarray manufacturers have realized the need for such focused arrays and have started to offer many of them. Typically, a focused array includes a few hundreds of genes covering the biological mechanism(s) being studied. However, two microarrays produced by different companies are extremely unlikely to use the same set of genes. In consequence, various pathways will be represented to various degrees on different arrays even if the arrays are all designed to investigate the same biological mechanisms. This is an unavoidable functional bias. Such a bias will be associated with each and all arrays that include less than the full genome of a given organism.
The Onto-Tools (OT) toolkit helps researchers assess the biological bias of various commercial arrays through its Onto-Compare (OC) tool. The Onto-Compare database is populated with data collected from several online databases, as well as the lists of genes (GenBank accession numbers) for each microarray as provided by their manufacturers. From the list of accession numbers, a list of unique UniGene cluster identifiers is prepared for each microarray, and then a list of LocusLink identifiers is created for each microarray from the list of UniGene cluster identifiers in the OC database. Each locus in the LocusLink database is annotated using ontologies from the Gene Ontology Consortium (http://www.geneontology.org) and ontologies from other researchers and companies. The Gene Ontology Consortium provides ontologies for biological processes, molecular functions and cellular components. The data from these databases and gene lists is parsed and entered into the Onto-Compare relational database. After creating a list of locus identifiers for each array, the list is used to generate the following profiles: biochemical functions, biological process, cellular role, cellular component and molecular function. The profiles for each microarray are stored in the database. The list of genes deposited on a microarray is static, but the annotations for those genes keep changing and are updated automatically, as more information becomes available.
Onto-Design
In many cases, researchers prefer to print their own arrays. One of the reasons for opting to print one's own custom array is that given the complexity of the biological research one may feel that none of the commercially available microarrays represent the targeted pathways and biological processes to the extent needed. Other reasons may be related to the dramatically reduced price of an in-house solution versus commercial arrays and the ability to adapt the arrays to one's own experimental design and use of controls. In order to design a microarray that constitutes a powerful and effective interrogation tool, a researcher has to choose genes that are representative of key mechanisms, pathways and biological processes. At present, the choice of genes to include on a certain microarray is a very laborious process requiring a high level of expertise. Furthermore, this process is very time consuming, even for experts, since they have to consult many online databases as well as perform an extensive literature review in order to find the set of genes that are involved in specific biological processes of interest. Onto-Design is a tool that is developed to assist in this gene selection process.
The OD interface allows the user to either upload a set of functional categories of interest (such as biological processes), or to browse through a graphical representation of a tree representing the Gene Ontology hierarchy. Actually the GO hierarchy is a directed acyclic graph (DAG), not a tree. The internal structure of the database represents correctly the GO but the interface is more conveniently represented as a tree. Categories linked through DAG links not contained in the tree are automatically travelled by the system in the appropriate way.
Onto-Translate
In the annotation world, the same piece of information can be stored and viewed differently across different databases. For instance, more than one Affymetrix probe identifier (ID) can refer to the same GenBank sequence (accession number) and more than one nucleotide sequence from GenBank can be grouped in a single UniGene cluster. The result of OE depends on whether the input list contains Affymetrix probe IDs, GenBank accession numbers or UniGene cluster IDs. In order to illustrate this, let us consider an input specified as a list of 10 Affymetrix probe ids. Let us assume that the results show that four out of 10 probes are involved in biological process A and the remaining six probes are involved in biological process B. Therefore the frequency of biological process A will be four and for the process B will be six. In order to interpret this, a researcher might need to use the data sheet provided with each Affymetrix array (or the NetAffy web site) to map these probe IDs into accession numbers. This reveals that the four probe IDs for the process A correspond to only two different accession numbers and the six probe IDs for the process B correspond to another two different accession numbers. Repeating the OE analysis using accession numbers will show that the frequency of both the processes A and B is two. Furthermore, mapping the accession number to UniGene cluster IDs shows that all four accession numbers actually come from the same UniGene cluster. Repeating the OE analysis using cluster IDs will show the frequency of both A and B as one.
This example illustrates that the user has to be aware of these relationships between the different forms of the data in order to interpret correctly the results. Furthermore, even if a user is aware of the relationships and knows how to convert them, most existing tools only allow conversions of individual genes. This makes the process of translating hundreds of genes absolutely unfeasible. Onto-Translate (OT) is a tool that allows the user to perform easily such translations of entire sets of genes. A user can input a list of genes specified by either Affymetrix probe IDs, GenBank accession numbers or UniGene cluster IDs, indicate the type of the list by clicking the appropriate radio button and request the translation on the input list in any of the remaining two forms by selecting the appropriate radio button for output list.
Tool integration
All tools in the Onto-Tools package use a consistent interface. Genes and functional categories can be prepared in advance and submitted to the tools as a text file with one entry per line. The results can generally be emailed back to the user.
As shown with the examples in the Results section, each of the Onto-Tools addresses a specific problem currently faced by microarrays users. However, the ensemble of the Onto Tools is more than the sum of its components. This has been achieved by seamlessly integrating the tools. For instance, it is possible to use a general purpose array such as Affymetrix HG133 in order to investigate a given condition by screening a large number of genes. The list of differentially regulated genes can be analyzed with OE in order to identify the functional categories that are relevant in the given condition. The user can inspect OE's results and select a smaller number of highly significant categories (see 4 for a discussion of the significance values associated with the OE analysis). Based on these highly significant categories, the researcher might formulate a hypothesis about the underlying biological phenomenon. At this point the user can seamlessly switch to ‘Onto-Compare’ in order to find and compare existing commercial arrays that might be useful in the testing of this specific hypothesis. If none of the commercially available arrays covers the necessary pathways to a satisfactory degree, the user can then switch to Onto-Design to create their own custom array representing the chosen biological processes.
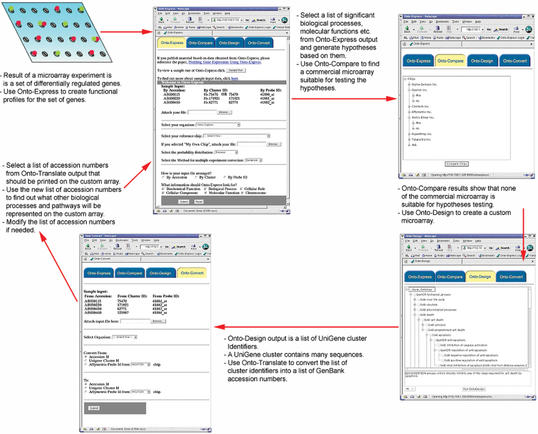
Results can also be seamlessly passed between Onto-Compare and Onto-Design. For example, the user compared all available commercial microarrays for apoptosis, was not satisfied with any of them and decided to create their own array. After designing a custom apoptosis array with Onto-Design, the user can click-switch back to Onto-Compare and compare the newly designed array with any of the existing commercial arrays. The user interfaces of the tools as well as a possible navigational pathway through the various tools are show in Figure 1. Future work will include an analysis at a specified level of the GO hierarchy.
Figure 1.
Onto-Tools (clockwise, from top left): regulated genes are analyzed with Onto-Express to find significantly impacted biological processes. Specific hypotheses can be formulated based on such processes. Onto-Compare can be used to select those commercial microarrays that cover best the hypothesized mechanism. If no satisfactory array is available, Onto-Design can be used to design a custom array for that specific set of hypotheses. Onto-Translate may be used at any time to translate between UniGene cluster IDs, accession numbers and array specific Affymetrix identifiers.
RESULTS
Onto-Express
OE's capabilities can be illustrated using an example from the literature (4). A microarray strategy was recently used to identify 231 genes (from an initial set of 25 000) that can be used as a predictor of clinical outcome for breast cancer (7). Using a classical approach based on putative gene functions and known pathways, Van't Veer et al. (7) identified several key mechanisms such as cell cycle, cell invasion, metastasis, angiogenesis and signal transduction as being implicated in cases of breast cancer with poor prognosis. The 231 genes found to be good predictors of poor prognosis were submitted to OE using the initial pool of 25 000 genes as the reference set. Our approach was validated by the fact that the results included most of the biological processes postulated to be associated with cancer including the positive control of cell proliferation, anti-apoptosis, oncogenesis, cell cycle control and cell growth and maintenance (Fig. 2). OE also identified a host of novel mechanisms. Protein phosphorylation was one of these additional categories significantly correlated with poor prognostic outcome. Apart from its involvement in a number of mitogenic response pathways, protein phosphorylation is a common regulatory tactic employed in cell cycle progression.
Figure 2.
Significant correlations were observed between the expression level and poor breast cancer outcome for 231 genes (7). This subset of genes was processed by Onto-Express to categorize the genes into functional groups. Red bar graphs represent genes for which the function was inferred, blue graphs represent genes for which the function was proved experimentally and green graphs represent genes for which this type of information was not recorded in the source database.
Onto-Compare
In order to illustrate the utility of Onto-Compare, we compared apoptosis arrays available from BD biosciences Clontech Inc. (Palo Alto, CA), Sigma–Genosys Inc. (Woodlands, Texas, US) and Perkin–Elmer Inc. (Wellesley, MA). Comparison of these three apoptosis specific arrays using Onto-Compare is shown in Table 1. Induction of apoptosis, tumor necrosis factor receptors and caspases are represented to a similar degree on all three arrays. Various interleukins are also reported to be mechanistically associated with apoptosis at both protein and gene levels (8–10). However, neither the Perkin–Elmer nor the Clontech microarray contains any interleukin related gene. On the other hand, the Sigma–Genosys microarray contains 14 such genes. Clearly, among the three arrays considered, the Sigma–Genosys would be a better choice to test any hypothesis involving the role of interleukins in apoptosis. Other processes such as immune response, cell–cell signalling, cell surface receptor linked signal transduction are also better represented on the Sigma–Genosys array. However, it is important to emphasize that the Sigma-Genosys array is not necessarily better than the other two arrays. In fact, since the Sigma–Genosys and the Clontech arrays have almost the same number of genes, there must exist some functional categories that are represented better on the Clontech array. Examples include processes such as cell cycle control, oncogenesis and negative control of cell proliferation.
Table 1. A comparison of three apoptosis specific microarrays: Clontech human apoptosis, Perkin–Elmer apoptosis and Sigma–Genosys human apoptosis.
| Ontology term | Clontech | Perkin–Elmer | Sigma–Genosys |
|---|---|---|---|
| Total genes on array | 214 | 346 | 210 |
| Induction of apoptosis by DNA damage | 3 (3) | 4 (4) | 3 (3) |
| Induction of apoptosis by extracellular signals | 8 (8) | 12 (12) | 7 (7) |
| Induction of apoptosis by intracellular signals | 2 (2) | 2 (2) | 2 (2) |
| Induction of apoptosis via death domain receptors | 4 (4) | 5 (5) | 7 (7) |
| Anti-apoptosis | 15 (15) | 20 (20) | 21 (21) |
| Regulation of cell cycle | 30 (30) | 30 (30) | 12 (12) |
| Induction of apoptosis | 16 (16) | 27 (26) | 23 (23) |
| Immune response | 0 (0) | 1 (1) | 19 (19) |
| Cell–cell signalling | 9 (9) | 9 (9) | 18 (18) |
| Oncogenesis | 22 (22) | 28 (28) | 16 (16) |
| Cell surface receptor linked signal transduction | 4 (4) | 9 (9) | 17 (17) |
| Positive regulation of cell proliferation | 5 (5) | 5 (5) | 12 (12) |
| Negative regulation of cell proliferation | 16 (16) | 20 (20) | 10 (10) |
| Caspases | 11 (10) | 16 (14) | 13 (13) |
| Tumor necrosis factor receptors | 4 (4) | 4 (4) | 4 (4) |
| Interleukins | 0 (0) | 0 (0) | 16 (16) |
| Unique sequences (Clusters) | 99 (98) | 133 (132) | 129 (129) |
While some categories (e.g. induction of apoptosis, caspases and tumor necrosis) are almost equally represented on each of the chips, some other are very unequally represented. For instance, neither Clontech nor Perkin–Elmer represent interleukins. Immune response, cell–cell signalling, cell surface receptor linked signal transduction are better represented on the Sigma–Genosys array. Other processes such as regulation of cell cycle, oncogenesis and negative regulation of cell proliferation are better represented on the Clontech array. The numbers represent sequences present on the arrays; the numbers in parentheses represent distinct UniGene clusters.
Onto-Design
In order to validate Onto-Design, we used it to design an array suitable for the study of apoptosis. We chose apoptosis because several such arrays are available commercially. We compared our self-designed custom array with three commercial arrays from Clontech (206 genes), Perkin–Elmer (324) and Sigma–Genosys (198). Our array has a similar size (250 genes) and yet it covers all biological processes relevant to apoptosis equally well or better compared to the three commercially available microarrays.
Our custom apoptosis was designed in two steps. In the first step, we selected all children of the apoptosis node from the biological processes tree of GO. In order to minimize the number of genes, OD was requested to provide a list of Unigene clusters not unique to a biological process. The results from OD are shown in the last column in Table 2. The results are shown in (first step)/(second step) format.
Table 2. Comparison of three commercial human apoptosis arrays with our custom designed apoptosis array.
| Biological process | Sigma–Genosys | Perkin–Elmer | Clontech | Custom array |
|---|---|---|---|---|
| Unique sequences(Clusters) | 210(198) | 346(324) | 214(206) | (229)/(250) |
| DNA fragmentation | 4(4) | 3(3) | 1(1) | (4) |
| DNA repair | 4(4) | 9(9) | 6(6) | (2) |
| I-kappaB phosphorylation | 2(2) | 0(0) | 0(0) | (0)/(2) |
| RAS protein signal transduction | 1(1) | 3(3) | 3(3) | (0)/(3) |
| Anti-apoptosis | 21(21) | 20(20) | 15(15) | (53)/(55) |
| Apoptosis | 16(16) | 24(24) | 15(15) | (79)/(85) |
| Apoptotic program | 7(7) | 7(7) | 8(7) | (8)/(9) |
| Caspase activation | 0(0) | 1(1) | 0(0) | (5) |
| Caspase activation via cytochrome c | 1(1) | 1(1) | 1(1) | (2) |
| Cell death | 0(0) | 1(1) | 0(0) | (2) |
| Cell motility | 6(6) | 8(7) | 4(4) | (3)/(4) |
| Cell proliferation | 20(20) | 19(19) | 21(21) | (16)/(21) |
| Cell–cell signalling | 18(18) | 9(9) | 9(9) | (24)/(25) |
| Development | 9(9) | 4(4) | 4(4) | (11)/(13) |
| Immune response | 19(19) | 1(1) | 0(0) | (9)/(10) |
| Induction of apoptosis | 23(23) | 27(26) | 16(16) | (53)/(56) |
| Induction of apoptosis by DNA damage | 3(3) | 4(4) | 3(3) | (5)/(6) |
| Induction of apoptosis by extracellular signals | 7(7) | 12(12) | 8(8) | (18) |
| Induction of apoptosis by hormones | 1(1) | 1(1) | 1(1) | (4) |
| Induction of apoptosis by intracellular signals | 2(2) | 2(2) | 2(2) | (7) |
| Induction of apoptosis by oxidative stress | 1(1) | 0(0) | 0(0) | (0)/(1) |
| Induction of apoptosis via death domain receptors | 7(7) | 5(5) | 4(4) | (8) |
| Inflammatory response | 8(8) | 4(4) | 2(2) | (9) |
| Killing transformed cells | 0(0) | 1(1) | 0(0) | (1) |
| Killing virus-infected cells | 0(0) | 1(1) | 0(0) | (1) |
| Negative regulation of survival gene products | 1(1) | 2(2) | 2(2) | (4) |
| Neurogenesis | 3(3) | 5(5) | 2(2) | (8) |
| Positive regulation of cell proliferation | 12(12) | 5(5) | 5(5) | (3)/(13) |
| Proteolysis and peptidolysis | 6(6) | 7(7) | 7(6) | (7)/(8) |
| Regulation of CDK activity | 4(4) | 17(17) | 16(16) | (2)/(9) |
| Regulation of cell cycle | 12(12) | 30(30) | 30(30) | (16)/(32) |
| Signal transduction | 56(56) | 62(60) | 42(42) | (55)/(57) |
The array is designed in two steps. The differences in the results of both steps are highlighted in bold letters and the changes in the number of Unigene clusters are shown in the last column, separated by ‘/’.
In the first step, OD returned a list of 229 unique UniGene clusters. The second step involved the addition of certain biological processes, that are known to be relevant to apoptosis. These biological processes are shown in bold letters in Table 2. OD returned a total of 700 UniGene clusters. In order to minimize the number of genes on the custom array designed, we used the Onto-Compare feature that allows the user to manipulate subsets of genes. Arbitrary intersections and unions can be calculated. We used this feature and found that there are nine Unigene clusters common between regulation of CDK activity and regulation of cell cycle and 10 clusters in common between regulation of cell proliferation and regulation of cell cycle. Also, there is no cluster common between these two sets of nine and 10 clusters. Hence, we have a total 19 clusters, of which three already exist in the previous set of 229 clusters. Adding the new 16 genes makes the total number of clusters on the custom chip 245. The three Unigene clusters for RAS protein signal transduction and two Unigene clusters for I-kappaB phosphorylation are only representative of their respective processes. Adding these five clusters to the list brought the total number of clusters on the custom array to 250. This was our final design.
Our custom apoptosis array with 250 UniGene clusters was compared with the apoptosis arrays from BD Biosciences Clontech Inc., Perkin–Elmer and Sigma–Genosys Inc. The results are shown Table 2, in the second half of the last column. The comparison clearly shows that the custom array provides equal or more number of genes for each of the apoptosis related biological process compared to any of the commercial arrays.
DISCUSSION
The Onto-Tools package includes an annotation database as well as a number of data mining and analysis tools. Independently of the methods used to select these genes, the common task faced by any researcher is to translate these lists of genes into a better understanding of the biological phenomena involved. Currently, this is done through a tedious combination of searches through the literature and a number of public databases. We developed OE as a novel tool able to automatically translate such lists of differentially regulated genes into functional profiles characterizing the impact of the condition studied. OE constructs functional profiles (using Gene Ontology terms) for the following categories: biochemical function, biological process, cellular role, cellular component, molecular function and chromosome location. Statistical significance values are calculated for each category.
In most cases, the exploratory phase in which thousands of genes are screened is followed by a more focused, hypothesis driven stage in which certain specific biological processes and pathways are thought to be involved. Since a single biological process can still involve hundreds of genes, microarrays are still the preferred approach as proven by the availability of focused arrays from several manufacturers. Since focused arrays from different manufacturers use different sets of genes, each array will represent any given regulatory pathway to a different extent. We argue that a functional analysis of the arrays available should be the most important criterion used in the array selection. We developed Onto-Compare as a tool that can provide this functionality, based on the GO nomenclature.
However, many times, the research can be focused on specific pathways or phenomena for which no commercial arrays are available. In such cases, researchers may choose to design their own arrays. One of the important issues in array design is the choice of the genes to be deposited on the array. In particular, it is important that the genes chosen represent abundantly the biological processes and pathways relevant to the condition studied. Onto-Design is a tool that was developed to assist in this gene selection process. In order to validate Onto-Design, we used it to design an array suitable for the study of apoptosis. Several such arrays are available commercially and we compared our self-designed custom array with three commercial arrays from ClonTech (206 genes), Perkin–Elmer (324) and Sigma–Genosys (198). Our array has a similar size (250 genes) and yet it covers equally well or better all biological processes relevant to apoptosis. This shows that our approach is able to produce small and yet very effective focused arrays. We also designed an array for the study of estrogen regulated genes for which no commercial arrays are available to our knowledge (data not shown).
Onto-Translate is a utility that allows a convenient translation between any type of gene identifiers (accession numbers, UniGene cluster identifiers and Affymetrix probe identifiers). All tools above are integrated allowing the user to seamlessly pass data from one tool to another. The Onto-Tools package is available online at http://vortex.cs.wayne.edu/Projects.html. Users protected by a firewall must open port 8080 on their firewall in order to access these tools.
Acknowledgments
ACKNOWLEDGEMENTS
This work has been supported by the NSF grant number NSF-0234806, USMRMC grant number DAMD17-03-2-0035, NIH grant numbers RO1-NS045207-01 and R21-EB000990-01, and Michigan Life Sciences Corridor grant number MLSC-27.
REFERENCES
- 1.Schuler G.D., Boguski,M., Stewart,E., Stein,L., Gyapay,G., Rice,K., White,R., Rodriguez-Tome,P., Aggarwal,A., Bajorek,E. et al. (1996) A gene map of the human genome. Science, 274, 540–546. [PubMed] [Google Scholar]
- 2.Schuler G.D. (1997) Pieces of puzzle: expressed sequence tags and the catalog of human genes. J. Mol. Med., 75, 694–698. [DOI] [PubMed] [Google Scholar]
- 3.Pruitt K.D. and Maglott,D.R. (2001) Refseq and locuslink: Ncbi gene-centered resources. Nucleic Acids Res., 30, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drăghici S., Khatri,P., Martins,R.P., Ostermeier,G.C. and Krawetz,S.A. (2003) Global functional profiling of gene expression. Genomics, 81, 98–104. [DOI] [PubMed] [Google Scholar]
- 5.Khatri P., Drăghici,S., Ostermeier,G.C. and Krawetz,S.A. (2002) Profiling gene expression using Onto-Express. Genomics, 79, 266–270. [DOI] [PubMed] [Google Scholar]
- 6.Man M.Z., Wang,Z. and Wang,Y. (2000) POWER SAGE: comparing statistical tests for SAGE experiments. Bioinformatics, 16, 953–959. [DOI] [PubMed] [Google Scholar]
- 7.van't Veer L.J., Dai,H., van de Vijver,M.J., He,Y.D. Hart,A.A.M., Mao,M., Peterse,H.L., van der Kooy,K., Marton,M.J., Witteveen,A.T. et al. (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature, 415, 530–536. [DOI] [PubMed] [Google Scholar]
- 8.Aqeilan R., Kedar,R., Ben-Yehudah,A. and Lorberboum-Galski,H. (2003) Mechanism of action of interleukin-2 (IL-2)-Bax, an apoptosis-inducing chimaeric protein targeted against cells expressing the IL-2 receptor. Biochem. J., 370, 129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hodge S., Hodge,G., Flower,R., Reynolds,P., Scicchitano,R. and Holmes,M. (2002) Up-regulation of production of tgf-beta and il-4 and down-regulation of il-6 by apoptotic human bronchial epithelial cells. Immunol. Cell. Biol., 80, 537–543. [DOI] [PubMed] [Google Scholar]
- 10.Schuhknecht S., Duensing,S., Dallmann,I., Grosse,J., Reitz,M. and Atzpodien,J. (2002) Interleukin-12 inhibits apoptosis in chronic lymphatic leukemia (cll) b cells. Cancer Biother. Radiopharm., 17, 495–499. [DOI] [PubMed] [Google Scholar]