Abstract
The DSSP program automatically assigns the secondary structure for each residue from the three-dimensional co-ordinates of a protein structure to one of eight states. However, discrete assignments are incomplete in that they cannot capture the continuum of thermal fluctuations. Therefore, DSSPcont (http://cubic.bioc.columbia.edu/services/DSSPcont) introduces a continuous assignment of secondary structure that replaces ‘static’ by ‘dynamic’ states. Technically, the continuum results from calculating weighted averages over 10 discrete DSSP assignments with different hydrogen bond thresholds. A DSSPcont assignment for a particular residue is a percentage likelihood of eight secondary structure states, derived from a weighted average of the ten DSSP assignments. The continuous assignments have two important features: (i) they reflect the structural variations due to thermal fluctuations as detected by NMR spectroscopy; and (ii) they reproduce the structural variation between many NMR models from one single model. Therefore, functionally important variation can be extracted from a single X-ray structure using the continuous assignment procedure.
From discrete to continuous secondary structure assignment.
The automatic assignment of protein secondary structure from three-dimensional co-ordinates of protein structures is an important and, in principle, a simple bioinformatics tool. Assignments are used to visualise structures, to speed up computationally expensive structural comparisons and to improve sequence searches. Secondary structure is more conserved than sequence information. Statistics about secondary structure occurrence can be incorporated into a profile used for homology searches (1,2). This can yield improved accuracy over standard search tools using sequence-based information alone (2–6). Hence, secondary structure assignments are important to assure the optimal yield of experimental structures and to cleverly select the targets for structural genomics. Although a conceptually simple task, the assignment of secondary structure is not always well defined (1). In fact, assignments vary between different NMR models of the same protein and between X-ray structures of homologues (7). Previously, we argued that such differences are not a problem of the assignment scheme, rather that they carry important information if adequately processed. Indeed, the variations between different NMR models correlate with thermal disorder (7). The DSSP program developed by Kabsch and Sander (8) identifies secondary structure as described by Pauling and colleagues (9,10) for three helix types and two extended sheet types. DSSP has become the standard in the field. DSSPcont constitutes a relatively straightforward extension of DSSP by adding continuous assignments (Fig. 1). Because the continuous assignment of secondary structure reproduces the observed variation between high-quality NMR models, it also correlates with mobility related to protein function (7). Thus, continuous secondary structure assignments can recognise conformational variations from a single X-ray structure and thereby may assist predictions of functionally important residues. More generally, it may help to pave the way to automatically generate valid hypotheses from protein structures. Finally, the continuous assignment appeared to describe ends of regular secondary structure segments (helices and strand) more accurately than discrete assignments. Often these caps carry important information about function and structure. Hence, the continuum may sharpen the tools that already profit from discrete assignments.
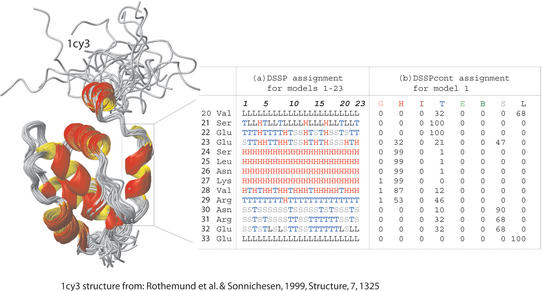
Figure 1.
DSSPcont assignment for 1c3y fragment. The variations between the secondary structure assignments for different NMR models of the same protein illustrate the impact of fluctuations on structure and highlight the difficulty of predicting protein structure. (A) The default DSSP assignments for all 23 models of the THP12-carrier protein [PDB identifier 1c3y (8)]. The structure models were calculated using 13C/15N labeled protein and 3D/4D NMR spectroscopy with 13 NOE's per residue. (B) DSSPcont assignments for the first NMR model alone.
Algorithm used to generate DSSPcont.
We assigned a continuum of secondary structure by running DSSP with nine different hydrogen bond thresholds (from −1.0 kcal/mol to −0.2 kcal/mol) (7). To score a given weighting scheme, we used the different models reported in NMR structure ensembles and calculated the average difference between single model assignments and the mean assignment. The best weighting scheme consequently ensured that the assignment extracted as much information as possible from a single NMR model given. The 100 best weighting schemes were all similar for helix {310-helix, alpha-helix, pi-helix}, strand {extended beta sheet, beta bridge} and other {other/loop, bend, helix-turn}. This similarity indicated that the weighting scheme had a well-defined stable global optimum. The most dominant weights were found close to the default DSSP hydrogen bond threshold of −0.5 kcal/mol. The weight for the −0.2 kcal/mol threshold was consistently low, while the adjacent threshold at −0.3 kcal/mol was consistently high. This prompted us to insert another threshold at −0.25 kcal/mol. To fine-tune the weighting scheme, we performed a simple gradient descent optimisation for 50, 100, 150 and 211 proteins. The DSSPcont assignment is therefore constructed by applying nine hydrogen bond thresholds from −0.2 kcal/mol in steps of 0.1 down to −1 kcal/mol, and in addition the tenth value of −0.25 kcal/mol. The result of the averaging procedure is that a single residue is no longer assigned a single ‘state’, rather the continuous secondary structure of a residue is characterised by a vector with propensities for the eight different DSSP states. More flexible residues have high propensities for more than one ‘state’, while ‘more frozen’ residues have non-zero values only for one particular state. This implies in particular that DSSPcont distinguishes well-defined from rigged helix/strand caps. Furthermore, DSSPcont distinguishes non-regular states that are flexible from those that are not.
Interface to web site.
The DSSPcont server can be accessed through a web interface for use on PDB formatted protein structures (http://cubic.bioc.columbia.edu/services/DSSPcont). Users may also access a DSSPcont database of pre-calculated assignments for all PDB (11) records; this database is updated weekly with all new PDB entries. The interface is very simple, requiring submission of a PDB identifier for the pre-calculated assignments. To run the DSSPcont algorithm on a user's own protein, a file containing the protein can be uploaded or the user can ‘cut and paste’ the protein description into the web interface. The DSSPcont predictions for all PDB entries have been integrated into the database integration system SRS (12). This enables to search by ‘ID’, ‘Compound Name’, ‘Source’, ‘Author Name’, ‘Number of Residues’, ‘Number of Chains’, ‘Total Number of Disulphide Bridges’, ‘Number of Intrachain Bridges’, ‘Interchain Disulphide Bridges’, ‘Protein Surface Accessibility’, ‘Total Number of Hydrogen Bonds’, ‘Number of Hydrogen Bonds in Parallel Bridges’ and ‘Hydrogen Bonds in Antiparallel Bridges’. The flat files for these DSSPcont assignments can be downloaded and used locally.
Output of DSSPcont.
The algorithm simply adds columns for the continuous assignment (as percentages for each of the eight states distinguished by DSSP) to the DSSP format (8). DSSP assigns eight states: 310-helix (represented by G), alpha-helix (H), pi-helix (I), helix-turn (T), extended beta sheet (E), beta bridge (B), bend (S) and other/loop (L). Eight columns are added to the standard DSSP output, each representing one of these DSSP states. In the example shown in Figure 1, there are 23 NMR models for the 1c3y fragment. Figure 1B shows the DSSPcont assignments for model 1. DSSP assigns the state of other/loop to residue 20 which is a valine. DSSPcont, however, is more detailed, predicting a 68% likelihood that it is involved in other/loop, but also a 32% probability of a helix turn. Each residue in the protein is assigned a percentage for each of the eight states. The core of the helix's residues (24–28) are assigned as H by default DSSP although the entire a-helix switched to a 310-helix when applying a hydrogen bond threshold of −1 kcal/mol. A ‘fuzzy’ helix capping, as seen here, is common and was observed for approximately one in four N-caps and half the C-caps in our data sets. Dissecting the continuous assignment shows that a 0.1 kcal/mol looser hydrogen bond threshold in the default DSSP would extend the helix by one residue (residue 29). If the default threshold instead had been tightened by 0.2 kcal/mol, the helix would lose one residue (residue 28). A more detailed online explanation of the DSSPcont format can be found at http://cubic.bioc.columbia.edu/services/DSSPcont/DSSPcont.html.
Acknowledgments
ACKNOWLEDGEMENTS
Thanks to Chris Sander (Sloan Kettering, New York) for the permission to use DSSP, to Gerrit Vriend (Nijmegen, Netherlands) for maintaining DSSP, to Arthur G. Palmer (Columbia University) and to Søren Brunak (Technical University of Denmark) for their invaluable contributions that were at the base of the scientific development of DSSPcont. Thanks to Jinfeng Liu (Columbia University) for computer assistance. The work was supported by the grants 1-P50-GM62413-01 and RO1-GM63029-01 from the National Institute of Health (NIH). Last, but not least, thanks to all those who deposit their experimental data in public databases and to those who maintain these databases.
REFERENCES
- 1.Andersen C.A.F. and Rost,B. (2003) Secondary structure assignment. In Bourne,P. and Weissig,H. (eds), Structural Bioinformatics. John Wiley, Hoboken, NJ, pp. 339–361. [Google Scholar]
- 2.Holm L. and Sander,C. (1999) Protein folds and families: sequence and structure alignments. Nucleic Acids Res., 27, 244–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jennings A.J., Edge,C.M. and Sternberg,M.J. (2001) An approach to improving multiple alignments of protein sequences using predicted secondary structure. Protein Eng., 14, 227–231. [DOI] [PubMed] [Google Scholar]
- 4.Rost B. (1995) TOPITS: threading one-dimensional predictions into three-dimensional structures. In Rawlings,C., Clark,D., Altman,R., Hunter,L., Lengauer,T. and Wodak,S. (eds), Third International Conference on Intelligent Systems for Molecular Biology. Menlo Park, CA: AAAI Press, Cambridge, England, pp. 314–321. [PubMed] [Google Scholar]
- 5.Rost B., Schneider,R. and Sander,C. (1997) Protein fold recognition by prediction-based threading. J. Mol. Biol., 270, 471–480. [DOI] [PubMed] [Google Scholar]
- 6.Fischer D. and Eisenberg,D. (1996) Fold recognition using sequence-derived properties. Protein Sci., 5, 947–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersen C.A., Palmer,A.G., Brunak,S. and Rost,B. (2002) Continuum secondary structure captures protein flexibility. Structure (Camb.), 10, 175–184. [DOI] [PubMed] [Google Scholar]
- 8.Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- 9.Pauling L., Corey,R.B. and Branson,H.R. (1951) Two hydrogen-bonded helical configurations of the polypeptide chain. Proc. Natl Acad. Sci. USA, 37, 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pauling L. and Corey,R.B. (1951) Configurations of polypeptide chains with favored orientations around single bonds: two new pleated sheets. Proc. Natl Acad. Sci. USA, 37, 720–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Etzold T. and Argos,P. (1993) SRS—an indexing and retrieval tool for flat file data libraries. Comput. Appl. Biosci., 9, 49–57. [DOI] [PubMed] [Google Scholar]