Abstract
Traditional treatment of infectious diseases is based on compounds that kill or inhibit growth of bacteria. A major concern with this approach is the frequent development of resistance to antibiotics. The discovery of communication systems (quorum sensing systems) regulating bacterial virulence has afforded a novel opportunity to control infectious bacteria without interfering with growth. Compounds that can override communication signals have been found in the marine environment. Using Pseudomonas aeruginosa PAO1 as an example of an opportunistic human pathogen, we show that a synthetic derivate of natural furanone compounds can act as a potent antagonist of bacterial quorum sensing. We employed GeneChip® microarray technology to identify furanone target genes and to map the quorum sensing regulon. The transcriptome analysis showed that the furanone drug specifically targeted quorum sensing systems and inhibited virulence factor expression. Application of the drug to P.aeruginosa biofilms increased bacterial susceptibility to tobramycin and SDS. In a mouse pulmonary infection model, the drug inhibited quorum sensing of the infecting bacteria and promoted their clearance by the mouse immune response.
Keywords: antagonists/biofilm/furanone/GeneChip/microarray/quorum sensing
Introduction
As the 21st century commences, it is becoming increasingly apparent that the 20th century, which opened with the promise of the eradication of most infectious diseases, closed with the specter of re-emergence of many deadly infectious diseases. It has become clear that bacteria can adapt by mutation to the selective pressure imposed by antibiotics. Today, a global concern has emerged that we are entering a post-antibiotic era with a reduced capability to combat microbes (Geddes, 2000).
A large number of opportunistic pathogenic Gram-negative bacteria employ N-acylhomoserine lactone (AHL) as their command language to coordinate population behavior during invasion and colonization of higher organisms (Hentzer et al., 2002a). The communication systems share common regulatory features: AHL signal molecules are synthesized from precursors by a synthase protein I and interact with transcriptional activator proteins R to regulate expression of target genes. AHL signaling is often referred to as quorum sensing (QS) because the system enables a given bacterial species to sense when a critical (i.e. quorate) population density has been reached in the host and in response activate expression of target genes required for succession (Fuqua et al., 1994).
QS-controlled genes often encode virulence factors and gene products required for bacteria–host interactions (Pirhonen et al., 1993; Parsek and Greenberg, 2000; Pearson et al., 2000). More importantly, there is growing evidence that QS influences more complex behavioral processes such as the ability to form surface-associated, structured and cooperative consortia referred to as biofilms (Davies et al., 1998; Eberl et al., 1999; Huber et al., 2001). Biofilm formation plays an important role in bacterial pathogenesis and is a common cause of persistent infections (Costerton et al., 1999; Høiby et al., 2001; Middleton et al., 2002; Singh et al., 2002). Biofilm bacteria are resistant to disinfectants, antibiotics and the action of host immune defenses (Koch and Høiby, 1993; Costerton et al., 1999).
Pseudomonas aeruginosa is an increasingly prevalent opportunistic human pathogen and is the most common Gram-negative bacterium found in nosocomial and life-threatening infections of immunocompromised patients (Van Delden and Iglewski, 1998). Patients with cystic fibrosis are especially disposed to P.aeruginosa infections, and for these persons the bacterium is responsible for high rates of morbidity and mortality (Høiby and Frederiksen, 2000; Lyczak et al., 2002).
Pseudomonas aeruginosa possesses two QS systems: the LasR–LasI and the RhlR–RhlI, with the cognate signal molecules N-(3-oxo-dodecanoyl)-l-homoserine lactone (OdDHL) and N-buturyl-l-homoserine lactone (BHL), respectively. The two QS circuits orchestrate a symphony of virulence factors such as exoproteases, siderophores, exotoxins and several secondary metabolites (Passador et al., 1993; Winson et al., 1995). In vitro immunoassays on human leukocytes have shown that OdDHL possesses immunomodulatory properties, for example, inhibition of lymphocyte proliferation and downregulation of tumor necrosis factor-α production and IL-12 production (Telford et al., 1998). In addition, OdDHL has been demonstrated to activate T cells in vivo to produce inflammatory cytokine γ-interferon (Smith et al., 2001) and thereby potentially promote a Th2-dominated response leading to increased tissue damage and inflammation.
We have attempted to attenuate bacterial pathogenesis by interfering with bacterial QS systems. Our approach is based on natural signal antagonists isolated from a marine environment. Seaweeds are devoid of advanced immune systems but some have evolved to rely, at least in part, on secondary metabolite chemistry for protection against colonizing organisms. In particular, the Australian red macro-alga (seaweed) Delisea pulchra is largely unfouled in nature due to the production of biologically active halogenated furanones (de Nys et al., 1993). These secondary metabolites are released at the surface of the plant at concentrations that inhibit colonization by both prokaryotes and eukaryotes (de Nys et al., 1995, 2002; Maximilien et al., 1998; Dworjanyn et al., 1999). We subsequently discovered that these compounds are QS inhibitors (QSIs), resulting in inhibition of colonization traits in a number of bacteria (Givskov et al., 1996; Gram et al., 1996; Maximilien et al., 1998; Hentzer et al., 2002b). The present article demonstrates that the P.aeruginosa communication systems can be blocked by a novel halogenated furanone compound. This is a highly specific and effective approach to attenuating bacterial virulence and controlling bacterial infections.
Results
Development of furanone compounds
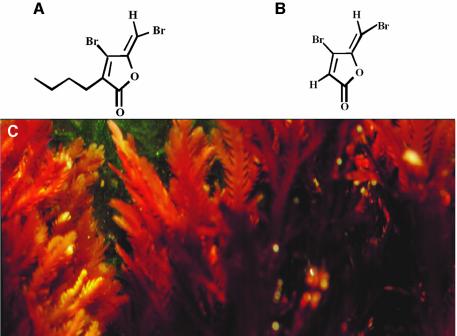
Our laboratories have previously reported on the generation of synthetic furanone compounds and their QSI activities (Manefield et al., 2002). In this work we have applied a novel substance, termed furanone C-30 (Figure 1). This compound displays an enhanced antagonistic activity against P.aeruginosa QS systems.
Fig. 1. From algal metabolite to Pseudomonas drug. (A) Compound 2, a natural furanone compound isolated from (C) D.pulchra. (B) compound C-30, a synthetic furanone with enhanced QSI activity.
Inhibition of virulence factor production
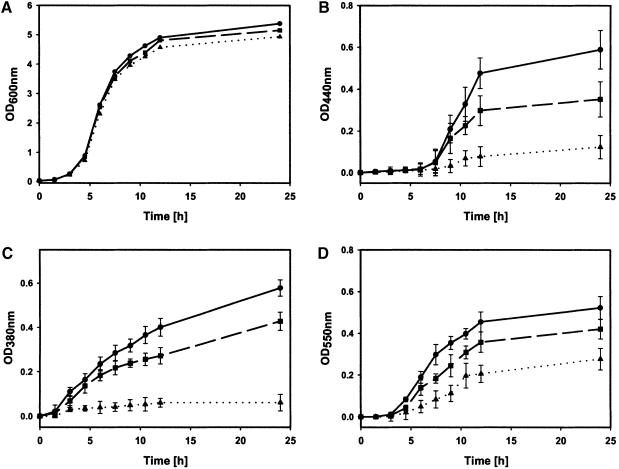
To test the efficacy of furanone C-30 to inhibit P.aeruginosa QS-controlled phenotypes, we investigated the effect on production of some QS-controlled extracellular virulence factors, namely protease, pyoverdin and chitinase. The production of these virulence factors was partially or completely suppressed in P.aeruginosa cultures grown in the presence of 1 or 10 µM (∼2.5 µg/ml) furanone C-30 (Figure 2). Importantly, the furanone did not affect growth of the planktonic cultures (Figure 2A). QS-deficient mutants of P.aeruginosa PAO1 show similar growth rates to the parental wild-type strain (Glessner et al., 1999).
Fig. 2. Influence of furanone C-30 on growth and expression of virulence factors of P.aeruginosa PAO1. Cultures were grown in the absence (circles) or presence of 1 µM (squares) and 10 µM (triangles) furanone C-30. (A) Growth rate; (B) exoprotease activity; (C) pyoverdin activity; (D) chitinase activity. The data represent mean values of three independent experiments. Error bars represent the standard errors of the means.
Identification of target genes of furanone C-30 action
QSI-screening assays and repression of P.aeruginosa virulence factor production suggest that QS circuits are targeted by the furanones. However, these observations do not exclude other targets of the furanone. DNA microarray technology offers the ability to overview the bacterial transcriptome and hence to reveal furanone target specificity by monitoring changes in transcript accumulation. We have used an Affymetrix GeneChip® P.aeruginosa Genome Array covering all the 5570 predicted P.aeruginosa PAO1 genes to study the effect of furanone C-30 on global gene expression.
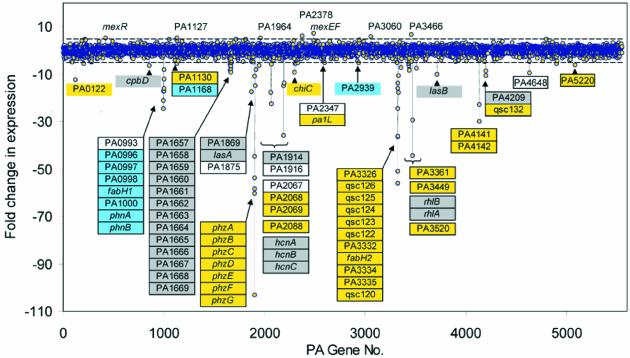
Planktonic cultures of P.aeruginosa PAO1 were grown with or without addition of furanone C-30. Analysis of microarray hybridization signals showed that 93 genes (1.7% of all genes) were >5-fold affected by the furanone compound (Figure 3). In all, 85 genes (1.5%) were repressed and eight genes (0.1%) were activated in response to C-30. About 43% of the C-30-regulated genes encode hypothetical proteins of unknown function. Currently, 44% of the P.aeruginosa genes are classified as encoding such hypothetical proteins (Whiteley et al., 2001). Among the 85 furanone-repressed genes, 30% have previously been reported as QS-controlled major P.aeruginosa virulence factors. These genes include the lasB gene encoding elastase, lasA encoding LasA protease, the rhlAB operon for rhamnolipid production, the phzA-G operon encoding phenazine biosynthesis, the hcnABC operon for hydrogen cyanide production and the chiC gene encoding chitinase activity.
Fig. 3. Effect of furanone C-30 on genome-wide gene expression profile of P.aeruginosa. Differential gene expression in planktonic cultures of P.aeruginosa PAO1 in response to 10 µM C-30 analyzed by microarrays. Positive values represent C-30-induced genes. Negative values indicate C-30-repressed genes. The dashed lines indicate 5-fold induction or repression. Genes significantly affected by C-30 are indicated. The color coding of the individual genes indicates which of the two QS systems are required for induction: blue, LasR-controlled genes (activated by addition of only OdDHL); gray, both LasR and RhlR are required for full expression (activated by OdDHL, and further induced by addition of BHL); yellow, RhlR-controlled genes (genes induced only by simultaneous addition of OdDHL and BHL). C-30-regulated genes recorded as non-QS-controlled are not colored. Data represent samples retrieved at an OD600 of 2.0.
Transcription of the lasRI and rlhRI genes was not significantly affected by C-30 (<2-fold repression). However, we observed that expression of two QS-associated genes, fabH1 and fabH2, was drastically repressed by C-30. These genes encode 3-oxo-acyl carrier protein (ACP) synthase III. Furthermore, two ACP-encoding genes (PA3334 and PA1869) were significantly downregulated by C-30. Acyl-ACPs have been proposed to be the acyl donors for synthesis of AHLs (Schaefer et al., 1996; Parsek et al., 1999).
Transcription of the phnAB operon encoding anthranilate synthase was repressed by C-30. Recently, the phnAB operon was suggested to encode the biosynthetic function for the Pseudomonas quinolone signal PQS (Gallagher et al., 2002). PQS signaling is, in concert with the AHL-based QS systems, involved in regulation of virulence factor production, in particular phenazine, pyocyanin and hydrogen cyanide, and in autolysis of P.aeruginosa colonies (D’Argenio et al., 2002), and hence potentially also in biofilms.
Among the C-30-activated genes we observed the mexEF genes encoding a multidrug efflux transporter and the mexR gene encoding the multidrug resistance operon repressor. Other C-30-activated genes included oxidoreductases, ABC transporters and MFS transporters.
To obtain an independent validation of our microarray data, we have studied expression of reporter fusions using the green fluorescent protein (GFP) as the reporter. We observed that reporter gene expression correlated well with microarray data. For example, GFP expression from a lasB promoter fusion was 7-fold repressed in the presence of 10 µM C-30 (data not shown). In comparison, microarray analysis showed that a similar C-30 treatment caused lasB mRNA accumulation to fall ∼10-fold. Virulence factor measurements (Figure 2) showed that exoprotease was reduced 9-fold in the presence of 10 µM C-30.
Mapping of the P.aeruginosa QS regulon
Among the 1.7% of P.aeruginosa genes that were significantly affected by C-30, one-third have previously been reported as QS controlled, many of which encode major P.aeruginosa virulence factors. Until recently, QS target genes have only been identified by genetic analysis, not by transcriptome analysis. As a consequence, a larger fraction of our C-30-repressed genes might be as yet unidentified QS target genes. We have performed an in-depth mapping of genes responsive to exogenous OdDHL and BHL using a lasI rhlI double mutant constructed from the sequenced PAO1 strain (Stover et al., 2000).
We identified 163 genes activated in response to addition of AHLs, corresponding to 2.9% of the PAO1 genome (Figure 4). The QS-induced genes are scattered throughout the genome, supporting the view that the QS systems function as global regulatory systems. Many of the identified QS-controlled genes are organized in putative operons. A simple estimate predicts 34 such operons. Sequence analysis showed that 20% of the QS-regulated genes and operons contained las box-like sequences in their corresponding promoter regions (Figure 4). Whiteley et al. (1999) categorized QS-controlled genes into four classes depending on early or late induction either by OdDHL or by OdDHL and BHL in combination. In our analysis we observed similar expression profiles notwithstanding different experimental conditions.
Fig. 4. Quorum-induced genes and C-30-repressed genes in P.aeruginosa. Quorum-induced genes identified in lasI rhlI mutant grown with or without added exogenous AHL signals. The transcription profile of the quorum-induced genes in P.aeruginosa PAO1 is shown together with the maximal repression caused by C-30. 1The gene number and name are from the Pseudomonas Genome Project (www.pseudomonas.com). Genes previously reported as QS regulated are shown in bold type. ], genes organized in operon. The criteria for organization into putative operons were as follows: (i) genes are transcribed in the same orientation; (ii) intergenic regions are <200 bp; (iii) genes exhibit similar transcription profiles. 2Description from the Pseudomonas genome project. qsc gene names refer to the study by Whiteley et al. (1999); pqs gene names refer to the study by Gallagher et al. (2002).
The induction profiles of AHL-controlled genes in the lasI rhlI double mutant are generally in good agreement with the profiles recorded in PAO1 (Figure 4). We observed that some QS-controlled genes were activated slightly later in the wild type than in the signal-complemented double mutant (e.g. the putative operon PA3327–PA3336), whereas other genes (e.g. PA2331) were activated earlier in the wild type than in the AHL-complemented mutant. The absolute expression level of quorum-induced genes is generally not lower in the wild-type strain despite the fact that AHL signals are produced and accumulate as a consequence of bacterial growth and are not present in high concentrations throughout the growth cycle as was the case with the double-mutant cultures. These observations indicate that the timing of QS-induced gene expression is only slightly altered in a signal-complemented lasI rhlI mutant.
Many genes show temporal induction patterns where they are not only activated in response to increasing cell density but also repressed in stationary phase, as illustrated by the expression profile of the putative operon covering PA4129–PA4130. Such observations suggest that additional regulators of QS are involved, but they remain to be characterized.
Comparative analysis of the C-30 target genes and the QS regulon shows that 80% of the furanone-repressed genes are also QS controlled, using a 5-fold cut-off limit for furanone repression and QS induction. Likewise, 46% of AHL-induced genes were >5-fold repressed by C-30, and another 39% were 2- to 5-fold repressed (Figure 4). In general, there is a strong correlation between genes strongly induced and repressed by AHLs and C-30, respectively. Quorum-induced genes that are <5-fold repressed by C-30 are generally 5-fold (or less) induced by exogenous AHLs. Importantly, C-30-repressed genes are found among all four classes of QS-controlled genes. A few QS-controlled genes fail to be efficiently repressed by furanone despite a strong AHL induction. In particular, rsaL, which encodes a putative regulator of QS, is highly (>1000-fold) expressed upon addition of AHLs to the lasI rhlI mutant culture, but rsaL transcription is not repressed by the addition of C-30.
In order to investigate which cellular processes are controlled by QS, we have categorized the genes into functional groups according to the annotation by PseudoCAP (Supplementary figure S2, available at The EMBO Journal Online). This analysis shows that P.aeruginosa QS-controlled genes are slightly overrepresented in functional groups related to virulence and survival during infection, for example, adaptation/protection, antibiotic resistance and susceptibility, central intermediary metabolism, fatty acid and phospholipid metabolism, and protein secretion/export apparatus. In particular, many of the secreted factors (21%, corresponding to 18 genes) produced by P.aeruginosa are under QS control. All genes involved in quinolone signal response are under QS control; however, this functional group consists of only seven genes organized in three transcriptional units. The functional classification of C-30-repressed genes is strikingly similar to the grouping of QS-controlled genes, which indicates that the furanone interferes with QS-controlled gene expression in an unbiased fashion.
We hypothesized that the constitution of the QS regulon might depend on experimental conditions. For instance, the biofilm environment might provide an ideal environment for QS signaling because bacteria are present in a very high local concentration. Additionally, the bacteria are believed to exhibit a biofilm-specific physiology radically different from that of bacteria in a planktonic mode of growth (Sauer et al., 2002). To test our hypothesis, we grew biofilms of P.aeruginosa PAO1 and the lasI rhlI mutant in a silicone tube biofilm reactor as described in the Supplementary data. Our analysis showed that 254 genes were AHL induced in P.aeruginosa biofilms. Since biofilms are heterogeneous populations consisting of cells in many different growth stages, we compared gene induction ratios in biofilm samples with the planktonic data averaged over all five cell-density sample points. Among the planktonic QS genes showing an average induction ratio >5-fold, 86% of them were also >5-fold induced in the biofilm samples (Supplementary table S1).
Effect on biofilm stress tolerance
Many commonly used traditional antibiotics have been demonstrated to be ineffective on biofilm cells compared with planktonic cells (Anwar and Costerton, 1990). We wanted to test whether C-30 shows a similar inadequacy to cope with biofilm bacteria.
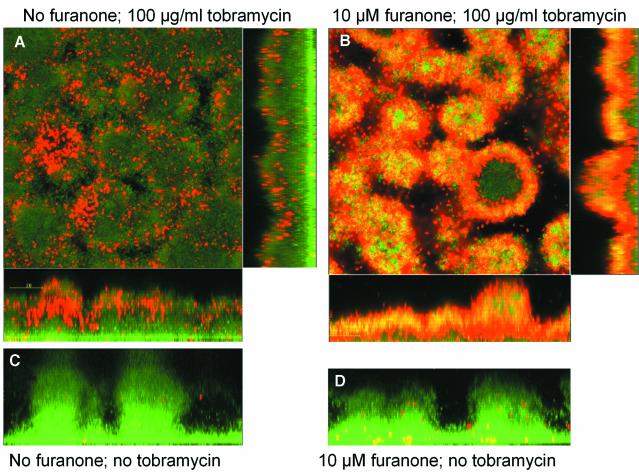
For P.aeruginosa, it has been demonstrated that the ability to form the characteristic mushroom-structured and SDS-resistant biofilms is affected by QS (Davies et al., 1998). We observed that biofilms grown in the presence of C-30 were, in contrast to a non-furanone-treated control, efficiently dissolved by an overnight treatment with 0.1% SDS (see Supplementary figure S1). The sensitivity to tobramycin, an aminoglycoside antibiotic routinely used in cystic fibrosis clinics (Høiby et al., 2000), was also assessed. Bacterial viability staining showed that biofilms grown in the presence of C-30 were significantly more susceptible to this antibiotic (Figure 5). The antibiotics efficiently penetrated and killed the furanone-treated biofilm cells, leaving 5–10% of cells alive (mainly present at the substratum). In the non-furanone-treated control, only the cells at the surface of the biofilm were killed by the tobramycin treatment. C-30-treated planktonic P.aeruginosa cells were two to three orders of magnitude more sensitive to tobramycin (data not shown).
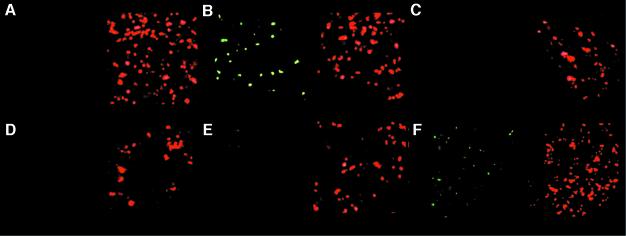
Fig. 5. Sensitivity of furanone C-30-treated P.aeruginosa biofilms to tobramycin. Scanning confocal laser microscopy (SCLM) photomicrographs of P.aeruginosa PAO1 biofilms grown in the absence (left panel) or presence (right panel) of 10 µM C-30. After 3 days, the biofilms were exposed to 100 µg/ml tobramycin for 24 h. Bacterial viability was assayed by staining using the LIVE/DEAD BacLight Bacterial Viability Kit: red areas are dead bacteria, and green areas are live bacteria. The biofilms were exposed to (A) no furanone and 100 µg/ml tobramycin, (B) 10 µM C-30 and 100 µg/ml tobramycin, (C) non-treated control and (D) 10 µM C-30 and no tobramycin.
QS inhibition in vivo
Cell-to-cell communication between infecting bacteria in the lungs of infected mice can be visualized by use of GFP reporter technology (Wu et al., 2000; Riedel et al., 2001). An Escherichia coli-based dual-labeled AHL sensor was introduced intratracheally to 4 × 107 colony-forming units (CFU) per lung. The sensor bacteria constitutively express the red fluorescent protein (RFP) for easy detection and localization in the lung tissue, and additionally express GFP in response to the presence of AHL signals. Introduction of OHHL into the mouse blood circulation caused activation of the LuxR-controlled PluxI-gfp(ASV) fusion (Figure 6A and B). This demonstrates that OHHL is transported by the blood, penetrates the lung tissue and induces QS-controlled gene expression in the infecting bacteria.
Fig. 6. Inhibition of QS in vivo. Photomicrographs of mouse lung tissue infected with an E.coli-based dual-labeled AHL sensor. The strain expresses GFP in response to the presence of exogenous AHL signals and carries a dsred expression cassette to provide a red fluorescent tag on the sensor bacteria for simple identification in tissue samples. Mice carrying the sensor bacteria in the lungs were administered OHHL and furanone C-30 via intravenous injection. (A) No injection; (B) 200 µM OHHL; (C) 200 µM OHHL + 2 µg/g BW C-30 (corresponds to ∼10 µM); (D) 400 µM OHHL + 2 µg/g BW C-30; (E) 800 µM OHHL + 2 µg/g BW C-30; (F) 1200 µM OHHL + 2 µg/g BW C-30.
We used this model system to evaluate the efficacy of C-30 in vivo. Furanone C-30 [∼2 µg/g body weight (BW)] co-administered intravenously with OHHL caused repression of LuxR-controlled activation of the dual-labeled AHL sensor (Figure 6C). C-30 inhibition was reversed by increasing dosages of OHHL (Figure 6D, E and F). This shows that the furanone compound can be transported by the blood circulation to the lungs, penetrate the lung tissues, enter the bacteria and, in turn, repress QS-controlled gene expression.
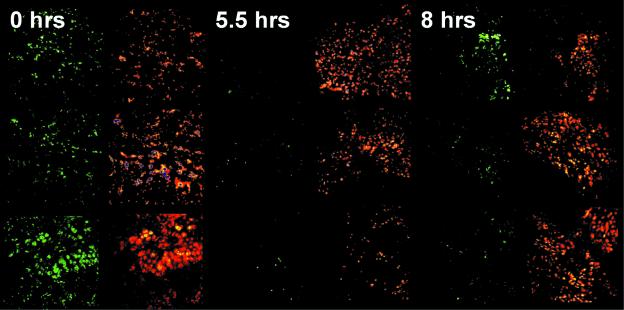
The ability to suppress P.aeruginosa QS in vivo was further tested by infecting mouse lungs with 2 × 107 CFU/lung of P.aeruginosa harboring a dual-labeled PA quorum sensor (Hentzer et al., 2002b). The infection was allowed to establish for 2 days before C-30 (∼1.7 µg/g BW) was introduced through the tail vein. Over a time span of 4–6 h after administration of C-30, the GFP signal from the dual-labeled quorum sensor was significantly reduced (Figure 7). After 8 h, the GFP signal reappeared, indicating that the furanone had cleared from the mouse blood circulation and hence de novo GFP synthesis recommenced (not shown). The experiments reveal important information about the mode of action of C-30. First, it significantly represses QS-regulated gene expression in vivo; secondly, the effect is concentration dependent; and thirdly, the compound is turned over within the duration of the experiment. This tells us that the QSI effect of a single C-30 injection lasts for ∼6 h in the present animal model.
Fig. 7. Inhibition of P.aeruginosa QS in mouse lungs. Photomicrographs of mouse lung tissue infected with P.aeruginosa carrying the dual-labeled PA quorum sensor for detection of QS signaling and a red fluorescent tag for simple identification in tissue samples. Mice were administered C-30 via intravenous injection at time zero. Infected animals were killed in groups of three at the time points indicated and the lung tissue samples were examined by SCLM.
Clearance of the infecting bacteria
Having established the conditions required for in vivo QS inhibition, the infection model was used to study the effect of C-30 on bacterial persistence in the lung. Twenty mice were infected with P.aeruginosa PAO1 at day 0. Immediately after this, the mice were split into two groups (10 each), which received subcutaneous injections of either C-30 (∼0.7 µg/g BW) or placebo with 8 h intervals for the following 3 days. Seven days post-infection, lungs were removed, homogenized and plated for CFU determination. The C-30-treated groups of animals displayed on average three orders of magnitude lower bacterial content than placebo groups (Figure 8). The efficiency of bacterial clearing was positively correlated with the concentration of C-30. In a similar experimental set-up, treatment with ∼0.4 µg/g BW resulted in a 10-fold lower CFU compared with the placebo group, whereas treatments with ∼0.2 µg/g BW had no detectable effect.
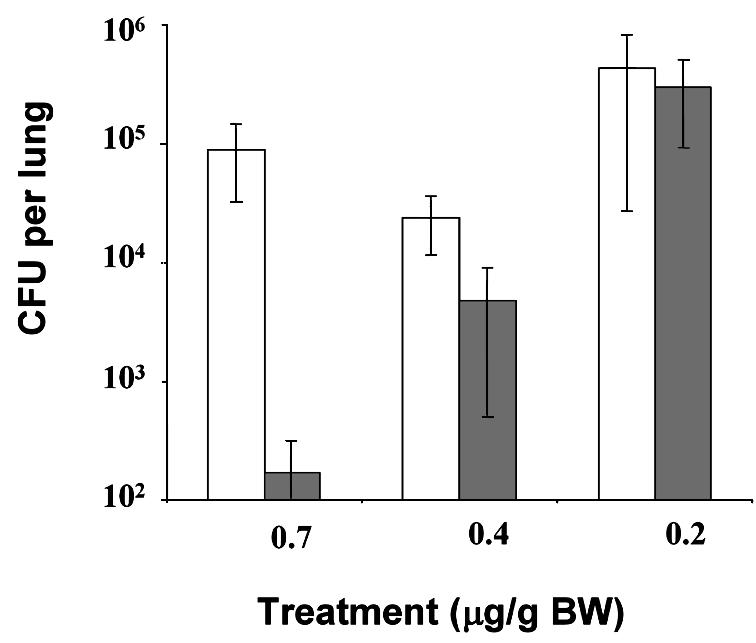
Fig. 8. Lung bacteriology. Healthy CBA/J mice (10 in each group) were infected with PAO1 as described in the text. Furanone C-30 (solid bars; concentration as indicated below the x-axis) or saline (open bars) were given three times a day (every 7 h) for 3 days. The mice were killed on day 7 after the bacterial challenge and the bacterial content of the infected lungs was determined as described in the text. Average values were plotted and the standard deviation is shown by error bars. The values were tested by means of an F-test (analysis of variance), and the p values for the 0.7, 0.4 and the 0.2 µM C-30 concentrations were 0.0007, 0.004 and 0.5, respectively.
Discussion
In this article we show that a synthetic halogenated furanone inhibits expression of QS-controlled behavior in P.aeruginosa, in particular the production of extracellular virulence factors, inhibits the development of antibiotic-resistant biofilms and reduces the persistence of infecting bacteria in a pulmonary mouse model. We have employed comprehensive molecular, genetic and biochemical techniques, as well as biological model systems, to support our conclusions.
Using microarray technology, we demonstrated that mRNA accumulation from 1.7% of the P.aeruginosa PAO1 genes is significantly affected by C-30. Among these, one-third have previously been reported as QS controlled, many of which encode major P.aeruginosa virulence factors. Among the C-30-repressed genes, we found many that have not previously been described as QS regulated. We decided to use transcriptome analysis to identify the QS regulon under the same conditions used to map the furanone target genes. We constructed a signaling-deficient lasI rhlI mutant from the sequenced P.aeruginosa PAO1 strain to identify furanone target genes and AHL-activated genes. Using a 5-fold cut-off, we identified 163 genes activated by quorum signals (Figure 4). Among these genes, most previously reported QS genes were present, though some, like toxA, sodA and kat, were absent. We examined the expression profiles of these genes in the wild type, but found that they were expressed below the detection limit. This shows, not surprisingly, that the constitution of the QS regulon depends on the experimental conditions.
We also compared the expression profiles of the AHL-induced genes in the signaling-deficient mutant with wild-type expression patterns. The expression profiles were similar despite differences in the experimental scenarios (mutant grown in the presence of saturating concentrations of AHLs). This observation is in agreement with those of other researchers, but nonetheless surprising in the light of the paradigm which states that QS-controlled induction occurs in response to build-up of extracellular signal molecules. The fact that gene expression cannot be induced prematurely at low cell densities in response to the addition of high doses of exogenous signals indicates that the onset of induction is not simply dictated by the signal concentration. Several other studies have pointed to regulatory factors involved in timing the onset of quorum induction; such factors include MvaT, QscR and RsmA (Chugani et al., 2001; Pessi et al., 2001; Diggle et al., 2002).
A comparative analysis of the QS regulon and C-30 target genes shows that 80% of the furanone-repressed genes are in fact controlled by QS. Furanone-repressed genes are not restricted to genes regulated by either the las or the rhl encoded systems, but are found throughout the continuum of QS-induced genes. Among the QS-induced genes, the majority were repressed by C-30. Importantly, the remaining non-repressed genes did not exhibit an obvious cell density-dependent expression profile in the wild-type strain. This is likely attributed to the different experimental conditions: QS-controlled genes were identified in a lasI rhlI mutant background grown with saturating levels of exogenous AHLs, whereas the furanone target genes were identified in the wild-type strain producing AHLs with increasing cell density. In essence, our analysis shows a clear overlap between strongly QS-induced genes and efficiently furanone-repressed genes. These genes include major virulence factors like lasA, lasB, hcnAB, rhlAB, chiC, phnAB and phzABCDEFG (see Figure 4). Intriguingly, expression of the lasI lasR and rhlI rhlR gene clusters, which encode the central components of the P.aeruginosa QS system, were not notably affected by the furanone treatment. This observation suggests that C-30 does not interfere with some of the regulatory systems controlling transcription of the lasRI and rhlRI genes, but rather that the furanone acts on these QS regulators at the post-transcriptional level.
Recently, two independent studies reported on identification of P.aeruginosa QS-controlled genes by microarray analysis (Schuster et al., 2003; Wagner et al., 2003). We have compared the results from the three studies in order to investigate whether the constitution of the QS regulon is affected by experimental factors. Our comparison shows a major overlap between the independent studies, but also points to several subsets of genes that appear to be QS controlled only under certain experimental conditions (Supplementary figure S3). We have termed the common group of QS-controlled genes the ‘general QS regulon’ of P.aeruginosa (Supplementary table S2). This designation relies entirely on data extracted from experiments using Affymetrix GeneChip® P.aeruginosa Genome Arrays and is sensitive to the significance cut-off value applied. Interestingly, the general QS regulon (except PA0144) was also induced by cells during biofilm growth (Supplementary table S1).
In a previous study, 39 QS-controlled genes were identified in P.aeruginosa by lacZ-based promoter probing (Whiteley et al., 1999). We have analyzed this data set as a control for our P.aeruginosa microarray data. The comparison shows good overall agreement between the independent data sets, both in terms of signal responses and signal specificities (Supplementary table S3).
Among the furanone target genes, we have identified the phnAB operon (covering PA0996–PA1002) (Cao et al., 2001). Similar data (Schuster et al., 2003; Wagner et al., 2003) show that this operon is subject to QS regulation. Previously, this operon was reported to be controlled by MvfR, a LysR-like transcriptional regulator required for maximum P.aeruginosa PA14 virulence in a plant leaf and the burned mouse model (Rahme et al., 2000) as well as in nematodes (Mahajan-Miklos et al., 1999). Recently, the phnAB operon was shown to encode enzymes responsible for biosynthesis of the Pseudomonas quinolone signal (PQS), thus giving rise to the alternative pqs operon designation (Gallagher et al., 2002). The PQS signaling system is hierarchically placed between the LasR and RhlR controlled circuits. Our array data confirm the proposed model in which pqsH is controlled by the las system (Gallagher et al., 2002), but in addition our analysis shows that the entire pqs operon is controlled by the las system. Interestingly, our data indicate that a las-dependent upregulation of mvfR expression precedes AHL-induced expression of the pqs operon.
The microarray analysis demonstrates that C-30 does not affect basal life processes. The furanones exhibit a high degree of specificity for the las and rhl quorum sensors. Furthermore, it is notable that C-30, in concentrations that significantly lower quorum-induced gene expression in planktonic cells, is equally active on biofilm bacteria despite the profoundly different lifestyles. In contrast, classical antibiotics used for the treatment of P.aeruginosa infections, such as tobramycin and piperacillin, are required in 100- to 1000-fold higher concentrations to eradicate biofilm bacteria compared with their planktonic counterparts (Anwar and Costerton, 1990). We propose that this difference is inherent to the furanone mode of action. Unlike classic antibiotics, which often target intracellular proteins involved in basal life processes, furanones might comprise a natural chemical defense system developed through the course of evolution to target and inactivate receptors of bacterial communication systems which, in turn, control virulence factor production, surface colonization and biofilm formation. Taken together, the effects on P.aeruginosa virulence factor production and biofilm sensitivity probably account for the rapid clearing of the bacteria in the pulmonary mouse model. Most importantly, the results demonstrate that bacterial virulence can be controlled by means of substances that specifically block cell-to-cell communication. Given the large number of bacteria that employ QS systems (Eberl, 1999), QSI compounds may find applications in many different settings, such as medicine, agriculture and food technology. Chemical attenuation of bacterial virulence, rather than bactericidal or bacteristatic strategies, is a highly attractive concept because such antipathogenic agents are less likely to pose a selective pressure for development of resistant mutants.
In the context of the present study, we envision that the concept of direct targeting virulence is promising as an early prophylactic treatment of individuals with P.aeruginosa infections. QSI drugs might prevent the formation of detrimental biofilms in the lung, on implants or in wounds. For cystic fibrosis patients, this might suffice to alter the delicate host–pathogen balance in favor of the host clearance mechanisms and thereby reduce the severity of infection.
Materials and methods
Bacterial strains
The P.aeruginosa PAO1 was obtained from the Pseudomonas Genetic Stock Center (www.pseudomonas.med.ecu.edu, strain PAO0001). This PAO1 isolate has served as the DNA source for the Pseudomonas Genome Project (www.pseudomonas.com) and subsequently as the template for the design of the P.aeruginosa GeneChip® (Affymetrix Inc., Santa Clara, CA). The lasI rhlI mutant was constructed using previously described knockout systems (Beatson et al., 2002). The knockout mutant was verified by genetic analysis and by screening for AHL production.
Detection of QS signaling
Detection of AHL signaling in planktonic cultures, biofilms and mouse lungs was achieved using an E.coli JM105 strain carrying the monitor plasmid pJBA132Gm, which encodes an unstable GFP reporter [luxR-PluxI-gfp(ASV)] for AHL detection (Wu et al., 2000; Andersen et al., 2001). For the present study, a dsred expression cassette was inserted into the EcoRV restriction site of this plasmid. This construct was referred to as a ‘dual-labeled AHL sensor’. OdDHL and BHL signaling were monitored using the P.aeruginosa PAO1 strain carrying a translational lasB-gfp(ASV) reporter fusion pMHLAS and a dsred expression cassette on a mini-Tn5 transposon element as described previously (Hentzer et al., 2002b). This is referred to as a ‘dual-labeled PA quorum sensor’.
Measurement of virulence factors
Strains were grown in Luria broth medium in the absence and presence of C-30 (1 and 10 µM) and harvested as described previously (Hentzer et al., 2002b). Assays for proteolytic activity and chitinase activity were performed as described elsewhere (Hentzer et al., 2002b). Pyoverdin production was monitored by 380 nm absorbance as described elsewhere (Prince et al., 1993).
Microarray analysis
Batch cultures of P.aeruginosa PAO1 were grown in ABt minimal medium with 0.5% casamino acids at 37°C and 200 r.p.m. A culture was inoculated with exponentially growing cells to an OD600 of 0.05. A reference sample for QS-controlled gene expression was retrieved at low cell density (OD600 = 0.5). The culture was split into two at a density of 0.7, and 10 µM furanone C-30 was added to one of the cultures. Further samples were retrieved at cell densities of 1.3, 1.6, 2.0 and 2.7. Samples for RNA isolation were immediately transferred to 2 vols of RNAlater (Ambion Inc., Austin, TX) and stored at –80°C. Microarray analysis of the QS regulon was performed by growing a culture of the lasI rhlI mutant to an OD600 of 0.3. The culture was split into three and the following additions were made: (i) no AHLs; (ii) 2 µM OdDHL; (iii) 2 µM OdDHL and 5 µM BHL. Samples were retrieved at the OD600 values given above. RNA purification and further processing for microarray analysis were performed according to the guidelines provided by the manufacturer of the GeneChip® P.aeruginosa Genome Array (Affymetrix Inc.). Microarray data analysis was performed using Affymetrix Microarray Suite 5.0 and Data Mining Tool 3.0 software. Average microarray hybridization signal intensity was scaled to 2500. For data analysis, the minimum and maximum signal thresholds were set to 200 and 100 000, respectively. The data represent the results of at least two independent experiments. A Mann–Whitney test was used to identify genes showing differential expression (p ≤ 0.05). In this article, we have only shown genes >5-fold repressed or induced and with an absolute difference in hybridization signal of >600. In our experimental settings, these criteria corresponded to a p value ≤ 0.03.
Biofilm experiments
Biofilms were cultivated in continuous-culture once-through flow chambers perfused with sterile ABtrace minimal medium containing 0.3 mM glucose as described previously (Hentzer et al., 2002b). Furanone C-30 was added to the ABtrace medium where appropriate. Biofilm development was examined by SCLM using a Zeiss LSM 510 system (Carl Zeiss GmbH, Jena, Germany) equipped with an argon laser and a helium–neon laser for excitation of fluorophores. Tolerance of biofilms to tobramycin and SDS was assessed by introducing 100 µg/ml tobramycin or 0.1% SDS, respectively, to the influent medium to 3-day-old biofilms. After 24 h, biofilms were examined by SCLM. Bacterial viability in planktonic and biofilm cultures was assessed using the LIVE/DEAD BacLight bacterial viability staining kit (Molecular Probes Inc., Eugene, OR) as described elsewhere (Hentzer et al., 2001).
Pulmonary mouse model
Healthy CBA/J mouse strains aged 11 weeks (average body weight 30 g) were used in animal studies. Immobilization of P.aeruginosa (5 × 108 CFU/ml) and E.coli (1 × 109 CFU/ml) in seaweed alginate beads and the surgical introduction of bacteria into mouse lungs (40 µl injected per lung) were performed as described previously (Wu et al., 2000). The mice were given OHHL by intravenous injection. The mouse total body volume was estimated to be 30 ml, which for the calculation of concentrations is ∼30 g. Furanone C-30 was administered by either intravenous or subcutaneous injection. AHL signal molecules and C-30 were prepared as stock solutions in 50% ethanol, which were diluted 20-fold in 0.9% NaCl prior to injection. Intravenous injection in the tail vein was applied to ensure that drugs were transported directly to the lung before passing the liver. Subcutaneous injections were applied where slow drug release was required. Preparation of lung tissue samples, subsequent SCLM examination and determination of lung bacteriology were performed as described elsewhere (Wu et al., 2000).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
This work was supported by grants from the Danish Technical Research Council, the Danish Medical Research Council, the Plasmid Foundation and the Villum Kann-Rasmussen Foundation to M.G., and from the Cystic Fibrosis Foundation, Therapeutics Inc. and the German Mukoviszidose e.V. to M.G. and L.E. M.A.S. was supported by a Skou stipend. The technical assistance of Linda Stabell was greatly appreciated.
References
- Andersen J.B., Heydorn,A., Hentzer,M., Eberl,L., Geisenberger,O., Christensen,B.B., Molin,S. and Givskov,M. (2001) gfp based N-acyl-homoserine-lactone monitors for detection of bacterial communication. Appl. Environ. Microbiol., 67, 575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar H. and Costerton,J.W. (1990) Enhanced activity of combination of tobramycin and piperacillin for eradication of sessile biofilm cells of Pseudomonas aeruginosa. Antimicrob. Agents Chemother., 34, 1666–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatson S.A., Whitchurch,C.B., Semmler,A.B.T. and Mattick,J.S. (2002) Quorum sensing is not required for twitching motility in Pseudomonas aeruginosa. J. Bacteriol., 184, 3598–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Krishnan,G., Goumnerov,B., Tsongalis,J., Tompkins,R. and Rahme,L.G. (2001) A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc. Natl Acad. Sci. USA, 98, 14613–14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugani S.A., Whiteley,M., Lee,K.M., D’Argenio,D., Manoil,C. and Greenberg,E.P. (2001) QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA, 98, 2752–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton J.W., Stewart,P.S. and Greenberg,E.P. (1999) Bacterial biofilms: a common cause of persistent infections. Science, 284, 1318–1322. [DOI] [PubMed] [Google Scholar]
- D’Argenio D.A., Calfee,M.W., Rainey,P.B. and Pesci,E.C. (2002) Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol., 184, 6481–6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies D.G., Parsek,M.R., Pearson,J.P., Iglewski,B.H., Costerton,J.W. and Greenberg,E.P. (1998) The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science, 280, 295–298. [DOI] [PubMed] [Google Scholar]
- de Nys R., Wright,A.D., König,G.M. and Sticher,O. (1993) New halogenated furanones from the marine alga Delisea pulchra. Tetrahedron, 49, 11213–11220. [Google Scholar]
- de Nys R., Steinberg,P.D., Willemsen,P., Dworjanyn,S.A., Gabelish,C.L. and King,R.J. (1995) Broad spectrum effects of secondary metabolites from the red alga Delisea pulchra in antifouling assays. Biofouling, 8, 259–271. [Google Scholar]
- de Nys R., Dworjanyn,S. and Steinberg,P.D. (2002) A new method for quantification of surface borne algal secondary metabolites. Mar. Ecol. Prog. Ser., 162, 79–87. [Google Scholar]
- Diggle S.P., Winzer,K., Lazdunski,A., Williams,P. and Camara,M. (2002) Advancing the quorum in Pseudomonas aeruginosa: MvaT and the regulation of N-acylhomoserine lactone production and virulence gene expression. J. Bacteriol., 184, 2576–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworjanyn S., de Nys,R. and Steinberg,P.D. (1999) Localization of secondary metabolites in the red alga Delisea pulchra. Mar. Biol., 133, 727–736. [Google Scholar]
- Eberl L. (1999) N-acyl homoserinelactone-mediated gene regulation in Gram-negative bacteria. Syst. Appl. Microbiol., 22, 493–506. [DOI] [PubMed] [Google Scholar]
- Eberl L., Molin,S. and Givskov,M. (1999) Surface motility of Serratia liquefaciens MG1. J. Bacteriol., 181, 1703–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua C., Winans,S.C. and Greenberg,E.P. (1994) Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol., 176, 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher L.A., McKnight,S.L., Kuznetsova,M.S., Pesci,E.C. and Manoil,C. (2002) Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol., 184, 6472–6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geddes A. (2000) Infection in the twenty-first century: Predictions and postulates. J. Antimicrob. Chemother., 46, 873–878. [DOI] [PubMed] [Google Scholar]
- Givskov M., de Nys,R., Manefield,M., Gram,L., Maximilien,R., Eberl,L., Molin,S., Steinberg,P.D. and Kjelleberg,S. (1996) Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J. Bacteriol., 178, 6618–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner A., Smith,R.S., Iglewski,B.H. and Robinson,J.B. (1999) Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of twitching motility. J. Bacteriol., 181, 1623–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gram L., de Nys,R., Maximilien,R., Givskov,M., Steinberg,P. and Kjelleberg,S. (1996) Inhibitory effects of furanones from the red alga Delisea pulchra on swarming motility of Proteus mirabilis. Appl. Environ. Microbiol., 62, 4284–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentzer M., Teitzel,G.M., Balzer,G.J., Heydorn,A., Molin,S., Givskov,M. and Parsek,M.R. (2001) Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol., 183, 5395–5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentzer M., Givskov,M. and Parsek,M.R. (2002a) Targeting quorum sensing for treatment of chronic bacterial biofilm infections. Lab. Med., 33, 295–306. [Google Scholar]
- Hentzer M. et al. (2002b) Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology, 148, 87–102. [DOI] [PubMed] [Google Scholar]
- Høiby N. and Frederiksen,B. (2000) Microbiology of cystic fibrosis. In Hodson,M.E. and Geddes,D.M. (eds), Cystic Fibrosis. Arnold, London, UK, pp. 83–107. [Google Scholar]
- Høiby N., Krogh,J.H., Moser,C., Song,Z., Ciofu,O. and Kharazmi,A. (2001) Pseudomonas aeruginosa and the in vitro and in vivo biofilm mode of growth. Microbes Infect., 3, 23–35. [DOI] [PubMed] [Google Scholar]
- Huber B., Riedel,K., Hentzer,M., Heydorn,A., Gotschlich,A., Givskov,M., Molin,S. and Eberl,L. (2001) The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology, 147, 2517–2528. [DOI] [PubMed] [Google Scholar]
- Koch C. and Høiby,N. (1993) Pathogenesis of cystic fibrosis. Lancet, 341, 1065–1069. [DOI] [PubMed] [Google Scholar]
- Lyczak J.B., Cannon,C.L. and Pier,G.B. (2002) Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev., 15, 194–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan-Miklos S., Tan,M.W., Rahme,L.G. and Ausubel,F.M. (1999) Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa–Caenorhabditis elegans pathogenesis model. Cell, 96, 47–56. [DOI] [PubMed] [Google Scholar]
- Manefield M., Rasmussen,T.B., Henzter,M. andersen,J.B., Steinberg,P., Kjelleberg,S. and Givskov,M. (2002) Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology, 148, 1119–1127. [DOI] [PubMed] [Google Scholar]
- Maximilien R., de Nys,R., Holmstrom,C., Gram,L., Givskov,M., Crass,K., Kjelleberg,S. and Steinberg,P. (1998) Chemical mediation of bacterial surface colonisation by sendary metabolites of the red alga Delisea pulchra. Aquat. Microb. Ecol., 15, 233–246. [Google Scholar]
- McKnight S.L., Iglewski,B.H. and Pesci,E.C. (2000) The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol., 182, 2702–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton B., Rodgers,H.C., Camara,M., Knox,A.J., Williams,P. and Hardman,A. (2002) Direct detection of N-acylhomoserine lactones in cystic fibrosis sputum. FEMS Microbiol. Lett., 207, 1–7. [DOI] [PubMed] [Google Scholar]
- Parsek M.R. and Greenberg,E.P. (2000) Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc. Natl Acad. Sci. USA, 97, 8789–8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek M.R., Val,D.L., Hanzelka,B.L., Cronan,J.E.J. and Greenberg,E.P. (1999) Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl Acad. Sci. USA, 96, 4360–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passador L., Cook,J.M., Gambello,M.J., Rust,L. and Iglewski,B.H. (1993) Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science, 260, 1127–1130. [DOI] [PubMed] [Google Scholar]
- Pearson J.P., Feldman,M., Iglewski,B.H. and Prince,A. (2000) Pseudomonas aeruginosa cell-to-cell signaling is required for virulence in a model of acute pulmonary infection. Infect. Immun., 68, 4331–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessi G., Williams,F., Hindle,Z., Heurlier,K., Holden,M.T., Camara,M., Haas,D. and Williams,P. (2001) The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol., 183, 6676–6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirhonen M., Flego,D., Heikinheimo,R. and Palva,E.T. (1993) A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J., 12, 2467–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince R.W., Cox,C.D. and Vasil,M.L. (1993) Coordinate regulation of siderophore and exotoxin A production: molecular cloning and sequencing of the Pseudomonas aeruginosa fur gene. J. Bacteriol., 175, 2589–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahme L.G. et al. (2000) Plants and animals share functionally common bacterial virulence factors. Proc. Natl Acad. Sci. USA, 97, 8815–8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel K., Hentzer,M., Geisenberger,O., Huber,B., Steidle,A., Wu,H., Høiby,N., Givskov,M. and Eberl,L. (2001) N-acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology, 147, 3249–3262. [DOI] [PubMed] [Google Scholar]
- Sauer K., Camper,A.K., Ehrlich,G.D., Costerton,J.W. and Davies,D.G. (2002) Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol., 184, 1140–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A.L., Val,D.L., Hanzelka,B.L., Cronan,J.E.,Jr and Greenberg, E.P. (1996) Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc. Natl Acad. Sci. USA, 93, 9505–9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Lostroh,C.P., Ogi,T. and Greenberg,E.P. (2003) Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol., 185, 2066–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P.K., Parsek,M.R., Greenberg,E.P. and Welsh,M.J. (2002) A component of innate immunity prevents bacterial biofilm development. Nature, 417, 552–555. [DOI] [PubMed] [Google Scholar]
- Smith R.S., Fedyk,E.R., Springer,T.A., Mukaida,N., Iglewski,B.H. and Phipps,R.P. (2001) IL-8 production in human lung fibroblasts and epithelial cells activated by the Pseudomonas autoinducer N-3-oxododecanoyl homoserine lactone is transcriptionally regulated by NF-κB and activator protein-2. J. Immunol., 167, 366–374. [DOI] [PubMed] [Google Scholar]
- Stover C.K. et al. (2000) Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature, 406, 959–964. [DOI] [PubMed] [Google Scholar]
- Telford G., Wheeler,D., Williams,P., Tomkins,P.T., Appleby,P., Sewell,H., Stewart,G.S., Bycroft,B.W. and Pritchard,D.I. (1998) The Pseudomonas aeruginosa quorum-sensing signal molecule N-(3-oxododecanoyl)-l-homoserine lactone has immunomodulatory activity. Infect. Immun., 66, 36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Delden C. and Iglewski,B.H. (1998) Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg. Infect. Dis., 4, 551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner V.E., Bushnell,D., Passador,L., Brooks,A.I. and Iglewski,B.H. (2003) Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol., 185, 2080–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley M., Lee,K.M. and Greenberg,E.P. (1999) Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA, 96, 13904–13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley M., Bangera,M.G., Bumgarner,R.E., Parsek,M.R., Teitzel,G.M., Lory,S. and Greenberg,E.P. (2001) Gene expression in Pseudomonas aeruginosa biofilms. Nature, 413, 860–864. [DOI] [PubMed] [Google Scholar]
- Winson M.K. et al. (1995) Multiple N-acyl-l-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA, 92, 9427–9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. et al. (2000) Detection of N-acylhomoserine lactones in lung tissues of mice infected with Pseudomonas aeruginosa. Microbiology, 146, 2481–2493. [DOI] [PubMed] [Google Scholar]