Abstract
The C to U editing of apolipoprotein B (apoB) mRNA is mediated by a minimal complex composed of an RNA-binding cytidine deaminase (APOBEC1) and a complementing specificity factor (ACF). This editing generates a premature termination codon and a truncated open reading frame. We demonstrate that the APOBEC1–ACF holoenzyme mediates a multifunctional cycle. The atypical APOBEC1 nuclear localization signal is involved in RNA binding and is used to import ACF into the nucleus as cargo. APOBEC1 alone induces nonsense-mediated decay (NMD). The APOBEC1–ACF complex edits and remains associated with the edited RNA to protect it from NMD. The APOBEC1 nuclear export signal is involved in the export of ACF and the edited apoB mRNA together, to the site of translation.
Keywords: ACF/APOBEC1/NMD/nucleocytoplasmic shuttling
Introduction
Cytidine (C) to uridine (U) RNA editing was first discovered in apolipoprotein B (apoB) mRNA where deamination of a C to U in mammalian apoB100 mRNA converts a glutamine codon (CAA) to a termination codon (UAA) midway along the 14 kb mRNA. The edited apoB mRNA is protected from nonsense-mediated decay (NMD) and apoB48 is generated which mediates the absorption of dietary lipid from the intestine. ApoB100 is made in the liver and transports endogenously synthesized cholesterol and triglyceride in the circulation (reviewed in Scott, 1995; Chester et al., 2000). Elevated levels of apoB100 are a major risk factor for coronary heart disease.
C to U editing of apoB mRNA is catalysed by a 27 kDa member of the cytidine deaminase family of enzymes, apoB mRNA editing catalytic polypeptide 1 (APOBEC1) (Navaratnam et al., 1993b; Teng et al., 1993). The C to U RNA editing deaminases also include recently identified proteins with homology to APOBEC1, namely APOBEC2 (Liao et al., 1999), activation-induced cytidine deaminase (AID) (Muramatsu et al., 1999), phorbolin-1 and phorbolin-like proteins (Madsen et al., 1999), and APOBEC3A–3G (Jarmuz et al., 2002). AID is involved in class switch recombination and hypermutation, APOBEC3A–3G are on chromosome 22 and expressed in a variety of cancer cell lines (Jarmuz et al., 2002). To date, no role in RNA editing has been demonstrated for any of these proteins. However, APOBEC3G is identical to CEM15, which has been shown to have anti-human immunodeficiency virus type 1 (HIV-1) activity (Sheehy et al., 2002).
Molecular modelling and extensive mutagenesis have shown that APOBEC1 is related in quaternary and tertiary structure to Escherichia coli cytidine deaminase (Navaratnam et al., 1998). Both enzymes form a homodimer with composite active sites constructed with contributions from each monomer. RNA binding by APOBEC1 is mediated through residues in and around its active site (Navaratnam et al., 1995). ApoB mRNA editing acquires its specificity from the primary sequence and secondary structure of the RNA (reviewed in Chester et al., 2000). The sequence around the editing site is AU rich and highly conserved from marsupials to man (Fujino et al., 1999). An 11 nucleotide sequence essential for specific editing (Shah et al., 1991), termed the mooring sequence, forms the downstream limb of this stem–loop structure (Richardson et al., 1998). Additional upstream elements have been identified that enhance editing activity (Hersberger and Innerarity, 1998), and have been incorporated into an alternative model (Gerber and Keller, 2001).
APOBEC1 alone is not sufficient for RNA editing. It requires APOBEC1 complementation factor (ACF/ASP) (Lellek et al., 2000; Mehta et al., 2000) for the editing of apoB mRNA. ACF has three non-identical RNA recognition motifs (RRMs) and belongs to the hnRNP R family of RNA-binding proteins. A splice variant of ACF, ACFΔ55, lacks 55 amino acids including the third RRM, and cannot complement editing (Chester et al., 2000). Deletion mutagenesis of ACF indicated that it binds to APOBEC1 through the third RRM and this region is involved in binding the RNA substrate (Blanc et al., 2001b; Mehta and Driscoll, 2002; N.Navaratnam, unpublished data).
Biochemical studies have shown that C to U editing of apoB mRNA is a nuclear event (Lau et al., 1991). APOBEC1 contains two runs of basic amino acids separated by a spacer region at the N-terminus which resembles a classic bipartite nuclear localization signal (NLS) (Dingwall and Laskey, 1991; Teng et al., 1993). Proteins that carry a basic NLS are recognized by the heterodimeric importin α/β receptor complex (Görlich and Mattaj, 1996). The crystal structure of yeast importin α has revealed two sites that can accommodate the NLS peptides within a helical surface groove (Conti et al., 1998).
APOBEC1 contains a C-terminal leucine-rich nuclear export signal (NES) (Yang et al., 1997). Both the NLS and NES in APOBEC1 have been implicated in RNA binding (Navaratnam et al., 1998). Nuclear transport signals not uncommonly overlap RNA-binding sites, and RNA binding can modulate transport events (La Casse and Lefebvre, 1995). One of the best characterized examples is the HIV-1 Rev protein, in which the bi-directional transport of Rev is mediated by an NLS and a leucine-rich NES (reviewed by Pollard and Malim, 1998). Several hnRNP proteins have also been shown to be shuttling proteins (Piñol-Roma and Dreyfuss, 1992; Flach et al., 1994); however, the steady-state localization of these proteins is predominantly nuclear.
Export of proteins from the nucleus utilizes various transport factors. Exportin 1 mediates the export of proteins with leucine-rich NESs (Fornerod et al., 1997). The antibiotic leptomycin B is a potent inhibitor of exportin 1-mediated nuclear export pathways and is used to dissect protein export processes (Wolff et al., 1997).
Normally, RNA containing a premature termination codon (PTC) is committed to degradation, eliminating mRNAs that would produce non-functional or harmful polypeptides so that only full-length proteins are expressed. The mechanism responsible for this has been termed NMD (Maquat, 1996; Hentze and Kulozik, 1999). An mRNA containing a PTC located at least 50 nucleotides upstream of the last exon–exon junction is recognized by the NMD surveillance machinery and committed to NMD (Nagy and Maquat, 1998; Lykke-Andersen et al., 2000). NMD in yeast takes place in the cytoplasm but, in mammalian cells, most NMD occurs in the cytoplasm with a fraction being nuclear (Frischmeyer and Dietz, 1999). It has been proposed that scanning of the mRNA occurs during its export from the nucleus or prior to export by a mechanism similar to cytoplasmic translation (Maquat and Carmichael, 2001). Indeed, it has been demonstrated that some translation occurs in the nucleus (Hentze, 2001; Iborra et al., 2001). There is an alternative model for NMD involving an exon junction complex (EJC), which is deposited 20 nucleotides upstream of exon–exon junctions and exported to the cytoplasm with the spliced mRNA (Le Hir et al., 2000). A physiological stop codon has no EJC downstream, but NMD-sensitive PTCs are followed by at least one EJC and this is proposed to mark them for NMD (Schell et al., 2002). Proteins required for NMD include the Upf family of proteins. Overexpression of an Arg844 to cysteine mutation of human Upf1 (R844C) was shown to exert a dominant-negative effect to prevent the decay of PTC-containing mRNAs (Sun et al., 1998).
In this study, we demonstrate that the APOBEC1–ACF complex performs multiple functions necessary to complete RNA editing and produce the truncated apoB protein, apoB48. Although some of the features of this cycle have been examined in isolation in other studies, here we have embellished these studies in the detail necessary to understand our novel data. We demonstrate that (i) APOBEC1 transports ACF to and from the nucleus as cargo; (ii) expression of APOBEC1 alone edits apoB RNA and generates a substrate for NMD; and (iii) the APOBEC1–ACF editing complex protects the edited apoB RNA from NMD and transports the RNA to the cytoplasm for translation.
Results
APOBEC1 has an atypical bipartite N-terminal NLS
Previous studies have demonstrated both nuclear and cytoplasmic localization of transfected APOBEC1 (Yang et al., 1997). Expression of endogenous APOBEC1 in cells and tissues is not high enough to allow immuno-detection (Yang et al., 1997; N.Navaratnam, unpublished data). Here we evaluated the intracellular localization of APOBEC1, expressed with a cytomegalovirus (CMV) promotor and detected by a C-terminal FLAG epitope tag. Transiently transfected APOBEC1 localized in the nucleus in all cells we examined, including those expressing ACF (CCL13, HepG2 and McA7777 cells), and lacking ACF (COS-7) (Figure 1A).
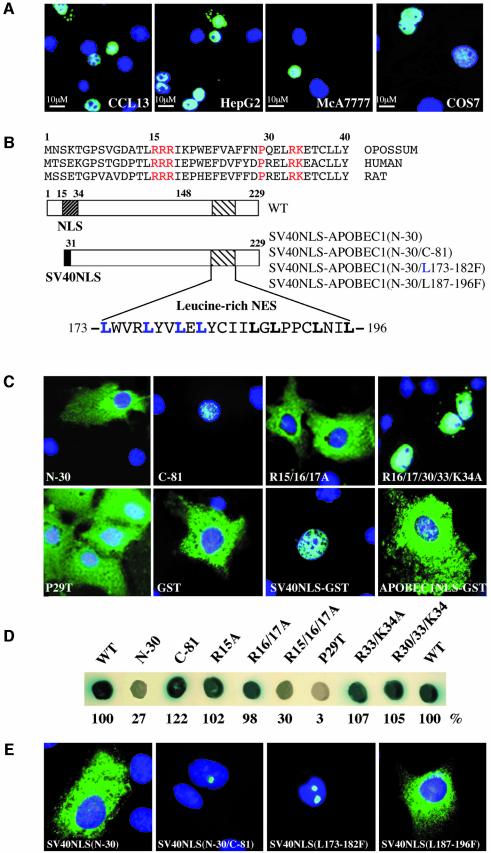
Fig. 1. APOBEC1 has a minimal bipartite N-terminal NLS and a leucine-rich NES. (A) Nuclear localization of C-terminal FLAG-tagged APOBEC1 in transiently transfected CCL13, HepG2, McA777 and COS-7 cells. The images show FITC overlaid with DAPI-stained nuclei. (B) Alignment of the N-terminal sequence of APOBEC1. The amino acid residues in the atypical bipartite NLS are shown in red and on a diagram of APOBEC1 (dark-hatched box) together with the leucine-rich NES (light-hatched box). The N-terminal 30 amino acids of APOBEC1 replaced with SV40NLS [SV40NLS–APOBEC1(N-30)], deletion of the C-terminal 81 amino acids of SV40NLS–APOBEC1(N-30) [SV40NLS–APOBEC1(N-30/C-81)] and leucines 173–182 or 187–196 mutated to phenylalanine [SV40NLS–APOBEC1(N-30/L173–182F or L187–197F)]. Leucines 173–182 are highlighted in blue, and 187–197 are shown in bold. (C) Intracellular localization of the APOBEC1 deletion (N-terminal deletion N-30, C-terminal deletion C-81) and combined mutants (R15/16/17A, R16/17/30/33/K34A) and point mutant (P29T) in COS-7 cells. Localization of GST, SV40NLS–GST and APOBEC1 NLS (1–44)–GST. The images show protein stained with FITC and overlaid with DAPI-stained nuclei. (D) Interaction of APOBEC1 and its mutants with importin α, analysed in the yeast two-hybrid system. Blue colonies denote interaction and white colonies denote no interaction. The strength of the interactions was also measured by liquid β-galactosidase assays and given as a percentage of the wild-type interaction. (E) Subcellular localization of the FITC images of SV40NLS–APOBEC1(N-30) and its mutant C-81, L173–182F and L187–196F proteins overlaid with DAPI.
The N-terminus of APOBEC1 contains an evolutionarily conserved (Fujino et al., 1999) basically charged, putative bipartite NLS, comprising arginine (R) amino acids 15–17, 30, 33 and lysine (K) 34, separated by proline (P) 29 (Figure 1B). To investigate this NLS, a series of deletion and point mutants of the NLS were constructed and transfected into COS-7 cells. Deletion of the N-terminal 30 amino acids from APOBEC1 (N-30) resulted in the mutant protein being completely excluded from the nucleus (Figure 1C). C-terminal deletion of 81 amino acids from APOBEC1 (C-81) resulted in nuclear accumulation (Figure 1C). To identify the critical residues in the NLS, alanine substitutions of the basic amino acids in the NLS cluster were created and the resulting proteins tested for their subcellular localization. Combined mutation of R15/16/17A resulted in cytoplasmic accumulation (Figure 1C). Mutation of R16/17/30/33A with K34A was nuclear. Mutation of P29 to threonine (P29T) abolished nuclear targeting and the protein was excluded from the nucleus (Figure 1C). Evaluation of the point mutations showed that at least any one of the arginine residues of the first cluster (R15–R17) and P29 are crucial for the nuclear localization of APOBEC1. These minimal requirements for nuclear localization are atypical for a bipartite NLS.
The NLS of APOBEC1 is not able or sufficient to target a heterologous cytoplasmic protein, GST, to the nucleus, whereas the SV40 large T antigen (SV40) NLS fused to GST directed it to the nucleus (Figure 1C). These results are consistent with previous findings of Yang et al. (1997).
Importin α is the nuclear import receptor for proteins containing an NLS with basic amino acids. A yeast two-hybrid assay was used to evaluate the interaction of importin α with the bipartite NLS of APOBEC1. A strong interaction was observed between APOBEC1 and importin α. The N-30 mutant of APOBEC1 showed much reduced interaction with importin α, whereas APOBEC1 with a C-terminal deletion of 81 amino acids (C-81) exhibited a wild-type level of interaction. Point mutations R15A/R16A/R17A and P29T that disrupted nuclear localization failed to interact with importin α, but mutations that did localize to the nucleus (R16A/R17A and R33A/K34A) interacted with importin α (Figure 1D). No interaction was observed between either importin β or transportin and APOBEC1, or with plasmid controls (data not shown). These data show that APOBEC1 interacts with importin α via its minimal NLS and indicate that it enters the nucleus by the importin α receptor-mediated pathway.
APOBEC1 has a leucine-rich NES
The C-terminus of APOBEC1 contains a 24 amino acid leucine-rich domain capable of overriding the SV40 NLS (Yang et al., 1997, 2001). This 24 amino acid domain encompasses two leucine-rich sequences between residues 173 and 196 (173–182 and 187–196) with homology to the NES found in HIV-1 Rev (Fischer et al., 1995) and in other leucine-rich NESs (Figure 1B). In order to refine the NES of APOBEC1, we assessed the subcellular localization of C-terminal deletions of the SV40NLS–APOBEC1(N-30) construct normally localized in the cytoplasm. Removal of 81 amino acids from the C-terminus of the 229 amino acid APOBEC1 protein (C-81) resulted in nuclear accumulation apparently within the nucleolus (Figure 1E); however, this was not confirmed by co-localization with known nucleolar proteins. To determine the minimal APOBEC1 NES, we divided the leucine residues into two groups (L173–L182 and L187–L196) and substituted them with phenylalanine (F) residues in the SV40NLS–APOBEC1(N-30) construct. Leucine mutations L173–L182F allowed nuclear targeting and remained in the nucleolus, whereas mutations L187–L196F localized in the cytoplasm (Figure 1E). Therefore, we conclude that leucine residues 173–182 are sufficient to export APOBEC1 from the nucleus.
APOBEC1 is a nucleocytoplasmic shuttling protein
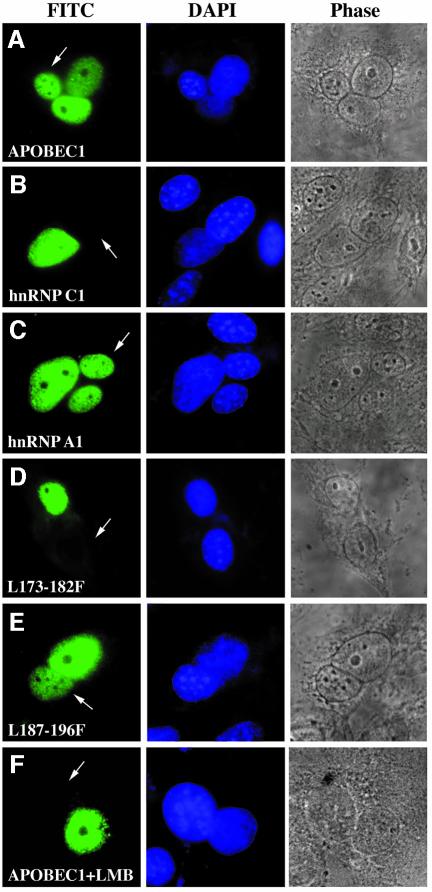
To examine the nucleocytoplasmic transport properties of APOBEC1, a transient, transfection interspecies heterokaryon assay was performed (Piñol-Roma and Dreyfuss, 1992). Human CCL13 cells transfected with APOBEC1 were fused to murine NIH-3T3 cells. Heterokaryons containing APOBEC1 displayed the protein in both nuclei (Figure 2A). In control experiments, the nuclear resident protein, hnRNP C1, was only found in the human nuclei, whilst the shuttling protein, hnRNP A1, was detected in both species of nuclei of the heterokaryons (Figure 2B and C). Thus, APOBEC1 is capable of shuttling between the nucleus and the cytoplasm.
Fig. 2. Exportin-mediated shuttling of APOBEC1 requires L173–L182. FLAG-tagged APOBEC1 wild-type (APOBEC1WT) (A), hnRNP C1 (B) and A1 (C) leucine mutants APOBEC1(L173–182F) (D) and APOBEC1(L187–196F) (E) were transfected into CCL13 cells and fused to mouse NIH-3T3 cells. (F) Heterokaryon of APOBEC1WT incubated with 50 ng/ml leptomycin B (LMB). Proteins were stained with anti-FLAG antibody and visualized with anti-mouse FITC (left). Nuclei were stained with DAPI (middle). Phase-contrast images of the heterokaryons are shown (right). The arrowhead indicates the mouse nuclei in the mouse–human heterokaryons.
To confirm the involvement of L173–L182 in the nuclear export of APOBEC1, the shuttling ability of APOBEC1 mutants was assessed. APOBEC1(L173–182F) (Figure 2D) and deletion mutant C-81 (data not shown) were only found in the CCL13 nuclei, whereas APOBEC1(L187–196F) was found in both nuclei (Figure 2E). These results further confirm that L173–L182 are sufficient for the nuclear export of APOBEC1.
The exportin 1-mediated nuclear export pathway acts through leucine-rich sequences (Fornerod et al., 1997) and is inhibited by leptomycin B (Wolff et al., 1997). The nucleocytoplasmic shuttling of APOBEC1 was completely blocked by leptomycin B (Figure 2F). HnRNP A1, which exits the nucleus via an exportin 1-independent pathway, (Fridell et al., 1997) was unaffected (data not shown).
APOBEC1 transports ACF to and from the nucleus as a cargo protein
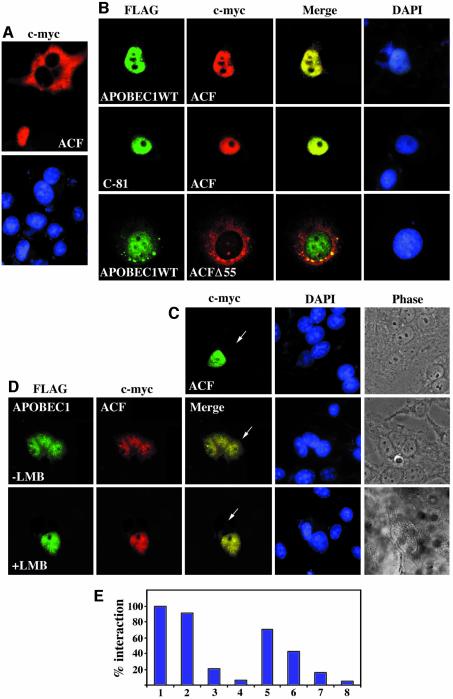
APOBEC1 is a nuclear protein (Figure 1A). ACF engineered with a C-terminal c-myc tag transiently transfected into CCL13 cells is localized predominantly in the cytoplasm (70%) but, in 30% of cells, ACF is nuclear (Figure 3A). Co-transfection of APOBEC1 with ACF localized both proteins predominantly to the nucleus (84%), with some cytoplasmic co-localization (16%) (Figure 3B). We therefore considered whether ACF might shuttle like APOBEC1. We evaluated nuclear ACF for shuttling by the interspecies heterokaryon assay. No shuttling was observed (Figure 3C). However, in the presence of APOBEC1, ACF co-localized with APOBEC1 in both the human and murine nuclei of the heterokaryon (Figure 3D). Co-shuttling of APOBEC1 and ACF was blocked by leptomycin B (Figure 3D). As expected, co-transfection of the nuclear resident APOBEC1 (C-81) with ACF co-localized to the nucleus (Figure 3B) but failed to exhibit shuttling in the heterokaryon assay (data not shown). ACF containing a 55 amino acid deletion (ACFΔ55) failed to co-localize to the nucleus with APOBEC1 (Figure 3B). This deletion spans the third RRM of ACF (RRM3) and is unable to interact with APOBEC1 (Blanc et al., 2001b; Mehta and Driscoll, 2002; N.Navaratnam, unpublished data). The nuclear import of ACF with APOBEC1 depends on the APOBEC1 NLS and the interaction between APOBEC1 and ACF RRM3, whereas export requires an intact APOBEC1 NES. The interaction between APOBEC1 and ACF allows the transport of ACF as cargo.
Fig. 3. APOBEC1 transports ACF as its cargo to and from the nucleus. (A) Subcellular localization of ACF c-myc (ACF) in CCL13 cells visualized with anti-c-myc-Cy3-conjugated antibody. (B) Subcellular localization of FLAG-tagged APOBEC1 wild-type (APOBEC1WT) or deletion of the C-terminal 81 amino acids of APOBEC1 (C-81), co-transfected with ACF c-myc (ACF) or APOBEC1WT with ACFΔ55 and visualized with anti-FLAG and anti-c-myc antibodies as before. Merged images show co-localization of ACF with APOBEC1WT and C-81 deletion mutant (top/middle), whereas APOBEC1WT with ACFΔ55 did not co-localize (bottom). DAPI-stained nuclei are also shown. (C) Heterokaryon analysis of nuclear ACF c-myc with anti-c-myc antibody and visualized with anti-mouse FITC. Nuclei were stained with DAPI, and a phase-contrast image of the heterokaryon is shown. (D) Co-transfected APOBEC1WT with ACF analysed by heterokaryon assay in the absence (top) and presence (bottom) of LMB. APOBEC1WT and ACF are visualized as before. Merged images show co-localization of both proteins in the mouse cells (arrowhead) in the absence of LMB. (E) Yeast two-hybrid analysis of the interaction of ACF with (1) APOBEC1, and its mutants (2) C-100, (3) N-15, (4) N-30, (5) R15/16A, (6) R15/16/17A, (7) P29T, and (8) vector control. The strength of the interaction measured by β-galactosidase assays is shown as a percentage of the wild-type interaction.
APOBEC1 C-81 enters the nucleus and can transport ACF into the nucleus (Figure 3B). This suggests that the site of interaction between APOBEC1 and ACF is located towards the N-terminus of APOBEC1. To map further the interaction sites on APOBEC1 and to establish whether this interaction involves the nuclear transport signals, we used the yeast two-hybrid system (Figure 3E). As previously demonstrated, the two full-length proteins exhibited a strong interaction (Blanc et al., 2001a). C-terminal deletion of 100 amino acids of APOBEC1 had no effect on this interaction. N-terminal deletion of 15 amino acids of APOBEC1 reduced the interaction with ACF to 24% compared with the wild-type interaction. N-30 deletion of APOBEC1 completely abolished this interaction. We therefore evaluated NLS mutants that disrupt nuclear localization. The level of interaction was unaffected by mutants that do not effect APOBEC1 nuclear localization (R16A/R17A). Interaction was decreased to 40% with APOBEC1 mutants R15A/R16A/R17A and almost abolished with P29T (<20%) (Figure 3E). The results indicate that ACF binds to APOBEC1 in the same region as the NLS.
APOBEC1 NLS and NES signals are involved in apoB RNA binding
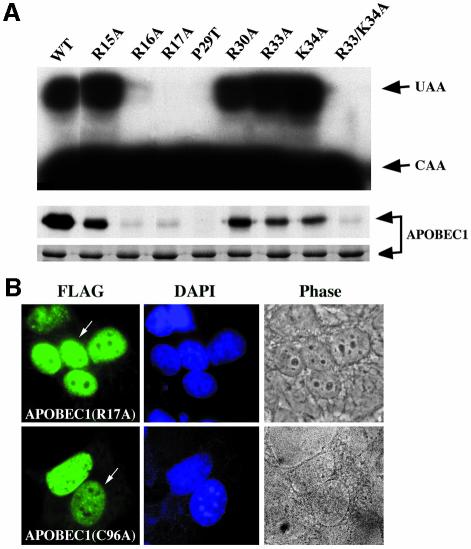
NLSs and NESs are known to be involved in RNA binding in several proteins (La Casse and Lefebvre, 1995). We considered the possible role of RNA binding in modulating the nuclear import and export of APOBEC1. In our previous studies, residues R16 and R17, which belong to the first cluster of basic amino acids in the NLS, were identified to be important for RNA binding and editing (Navaratnam et al., 1998). To evaluate a possible role for RNA binding of the other basic residues in the NLS, point mutations R33A and K34A and a combined mutant R33/K34A were examined. Mutants R33A and K34A behaved like wild-type, whereas the combined mutant abolished RNA binding and editing (Figure 4A). Homodimerization, a measure of APOBEC1 protein functional integrity, was unaffected by these mutations (data not shown). Mutant P29T, which abolished nuclear localization, behaved similarly to the combined mutant R33/K34A. Therefore, the basic amino acids and P29 within the bipartite NLS of APOBEC1 are crucial for nuclear localization and catalytic activity. Previously we have shown that L173 and L180 in the NES are involved in RNA binding and editing, but L177 and L182 are not (Navaratnam et al., 1998).
Fig. 4. APOBEC1 NLS and NES are involved in RNA binding. (A) RNA editing (top) and RNA UV cross-linking (middle) properties of GST fusion proteins of APOBEC1WT, point mutants R15, 16, 17, 30 33A, P29T and K34A and combined mutant R33/K34A. Edited (UAA), unedited (CAA) (top), RNA UV cross-linked and Coomassie-stained proteins (bottom) are shown. (B) Heterokaryon analysis of FLAG-tagged APOBEC1 mutants (R17A and C96A) visualized as in Figure 3.
To address whether nucleocytoplasmic shuttling of APOBEC1 requires apoB RNA binding, two APOBEC1 mutants, R17A, an NLS mutant unable to bind RNA, and C96A, an active site mutant with reduced RNA-binding ability (Navaratnam et al., 1998), were assessed for their ability to shuttle. Both mutants retained the ability to shuttle (Figure 4B), indicating that the nucleocytoplasmic shuttling of APOBEC1 can occur independently from apoB RNA binding.
C to U RNA editing by APOBEC1 generates a substrate for NMD
We considered why APOBEC1 and ACF need to shuttle in complex between the nucleus and cytoplasm. The editing of apoB mRNA is a co- or post-transcriptional event and generates a PTC. The introduction of a PTC in RNA generally designates it for NMD. The edited apoB RNA does not undergo NMD. We considered whether the assembly of the APOBEC1 and/or ACF complex on the apoB RNA might protect the edited RNA from NMD.
First, we verified that CCL13 cells are capable of executing NMD. The wild-type β-globin gene and the same gene containing a naturally occurring nonsense mutation at position 39 (PTC 39), previously used to evaluate NMD in HeLa cells (Thermann et al., 1998), were transfected into CCL13 cells. As expected, PTC 39 underwent NMD (Figure 5A).
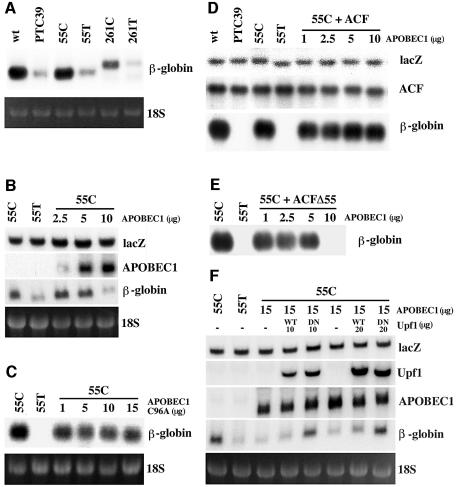
Fig. 5. ACF protects apoB RNA from APOBEC1-induced NMD. (A) Northern blot analysis of total RNA from CCL13 cells transfected with 10 µg of plasmid DNA containing β-globin wild-type (wt), PTC 39, 55 or 261 nucleotides of unedited (C) and edited (T) apoB RNA inserted in-frame in exon 2 of β-globin wild type (55C, 55T, 261C and 261T) in the same position as PTC 39. Northern blots probed with β-globin cDNA. Ethidium bromide staining of the 18S RNA from the corresponding gel is shown. (B) Northern blot analysis of total RNA obtained from cells transfected with plasmids 55T or 55C (10 µg) together with 5 µg of lacZ and 0–10 µg of APOBEC1 in a total of 30 µg of DNA per transfection and probed with lacZ, APOBEC1 and β-globin cDNAs. In some experiments, empty vector plasmid DNA was used to make up the total concentration of DNA to 30 µg. (C) Northern blot analysis of total RNA as in (B) except APOBEC1 was replaced with APOBEC1(C96A) catalytically inactive mutant and probed with β-globin cDNA. Other probes are not shown. (D) Northern blot analysis of total RNA from cells transfected with β-globin wild-type (wt), PTC 39, 55T or 55C (10 µg) together with 5 µg of lacZ, 30 µg of ACF and 1–10 µg of APOBEC1 in a total of 55 µg of DNA per transfection, and probed with lacZ, ACF and β-globin cDNAs. Empty vector plasmid DNA was used to make up the total concentration of DNA to 55 µg. (E) Northern blot analysis of total RNA as in (D) except ACF was replaced with ACFΔ55 and probed as in (D). Only the β-globin probe is shown. (F) Northern blot analysis of total RNA from cells transfected with 55T or 55C (10 µg) together with 5 µg of lacZ, 15 µg of APOBEC1 and 10–20 µg of Upf1 wild-type (WT) or R844C dominant-negative mutant (DN) in a total of 50 µg of DNA per transfection and probed with lacZ, Upf1, APOBEC1 and β-globin cDNAs. Empty vector plasmid DNA was used to make up the total concentration of DNA to 50 µg.
To evaluate whether apoB mRNA is sensitive to NMD, a 55 or 261 nucleotide apoB mRNA sequence, containing either the edited (55T/261T) or unedited (55C/261C) forms, was placed in-frame in exon 2 in the wild-type β-globin mRNA construct used above, in the same position as PTC 39. In transfected CCL13 cells, RNAs containing 55T and 261T nucleotides both underwent NMD similar to PTC 39. The 55C/261C RNAs were unaffected (Figure 5A). This process appears to be independent of the length of the apoB RNA sequence used; further experiments were performed with the 55C construct using 55T as a NMD control. Therefore, the edited apoB mRNA is sensitive to NMD, similar to what is seen in PTC 39.
Normally, edited apoB mRNA is protected from NMD, so we considered whether APOBEC1 might edit, remain bound to the RNA and protect the message from the NMD machinery. This was evaluated by co-transfecting increasing amounts of APOBEC1 with the 55C construct. Surprisingly, the presence of APOBEC1 led to the decay of 55C RNA in a dose-dependent manner compared with the control without APOBEC1 (Figure 5B). This may be due to the generation of multiple PTCs produced by APOBEC1 hyper-editing, similar to that described in transgenic mice overexpressing APOBEC1 (Yamanaka et al., 1997). To verify the induction of NMD by APOBEC1, we transfected APOBEC1 (C96A), a catalytically inactive mutant (Navaratnam et al., 1995). This mutant protein failed to generate a substrate for NMD (Figure 5C). Therefore, APOBEC1 alone cannot protect the edited apoB mRNA from undergoing NMD, rather it increased the RNA instability by presumably generating multiple PTCs by hyper-editing.
ACF protects apoB mRNA against NMD
ACF is essential to reconstitute C to U RNA editing by APOBEC1. Next, we considered whether the APOBEC1–ACF complex might protect the edited apoB RNA from decay. To evaluate the role of ACF, we transfected increasing amounts of APOBEC1 into CCL13 cells together with a fixed amount of ACF and the 55C construct used previously. In the absence of APOBEC1, ACF had no effect on 55C RNA and cannot protect 55T RNA or PTC 39 from NMD (Figure 5D). However, ACF and APOBEC1 together completely protected the edited 55C RNA from NMD (Figure 5D), even at the highest concentrations of APOBEC1 that cause NMD (Figure 5B). Under our experimental conditions, endogenous ACF was not detected by northern analysis. We also evaluated the splice variant of ACF (ACFΔ55) (Chester et al., 2000) that does not bind apoB RNA, interact with APOBEC1 or complement editing (Mehta and Driscoll, 2002; N.Navaratnam, unpublished data), and it failed to protect against NMD (Figure 5E). Therefore, we conclude that the APOBEC1–ACF interaction prevents hyper-editing by APOBEC1 and can protect the edited apoB RNA from undergoing NMD. RNA binding by ACF is important for the prevention of NMD, and this was supported further by the identification of apoB RNA in the APOBEC1–ACF complex (see below). This suggests that the role of ACF is to confer specificity for the C to U editing, to prevent hyper-editing and NMD of the apoB mRNA.
In order to verify that the APOBEC1-induced RNA decay occurs by the mechanism of NMD previously proposed, human Upf1 and its dominant-negative mutant (R844C) were transfected with β-globin-55C and APOBEC1. Mutant Upf1 protected against NMD in a concentration-dependent manner (Figure 5F). This suggests that the mechanism involved is similar to that which normally deletes PTC-containing mRNAs (Sun et al., 1998; Lykke-Andersen, 2000; Schell et al., 2002). However, a direct role for APOBEC1 in NMD cannot be ruled out.
The APOBEC1–ACF holoenzyme complex contains edited apoB mRNA
To establish whether the edited apoB RNA remains associated with the APOBEC1–ACF holoenzyme complex during nuclear export, nuclear and cytoplasmic extracts were prepared from transiently transfected CCL13 cells. The cells were transfected with β-globin-55C plasmid DNA together with ACF and/or APOBEC1 with c-myc and FLAG tags, respectively. The nuclear and cytoplasmic extracts were immunoprecipitated with the anti-c-myc or anti-FLAG antibodies. RNA isolated from the immunoprecipitates was analysed by semi-quantitative RT–PCR using β-globlin gene-specific primers that do not discriminate between edited or unedited RNA and the sequence verified.
RNA associated with ACF alone was completely unedited, whereas the level of editing in the APOBEC1–ACF-associated RNA was 50% in the cytoplasm and ∼10% in the nucleus (Figure 6A). Immunoprecipitation of ACF from cytoplasmic extracts recovered 50–90% of the input RNA (Figure 6B). Western blot analysis of extracts from cells transfected with APOBEC1 alone showed APOBEC1 present only in the nucleus, whereas in APOBEC1–ACF co-transfected cells, APOBEC1 was present in both the nucleus and cytoplasm. In the same extracts, ACF was also observed in both fractions (Figure 6A). These results indicate that the APOBEC1–ACF holoenzyme complex is involved in the editing and transport of the apoB RNA from the nucleus, as well as protecting it from NMD (see Figure 7).
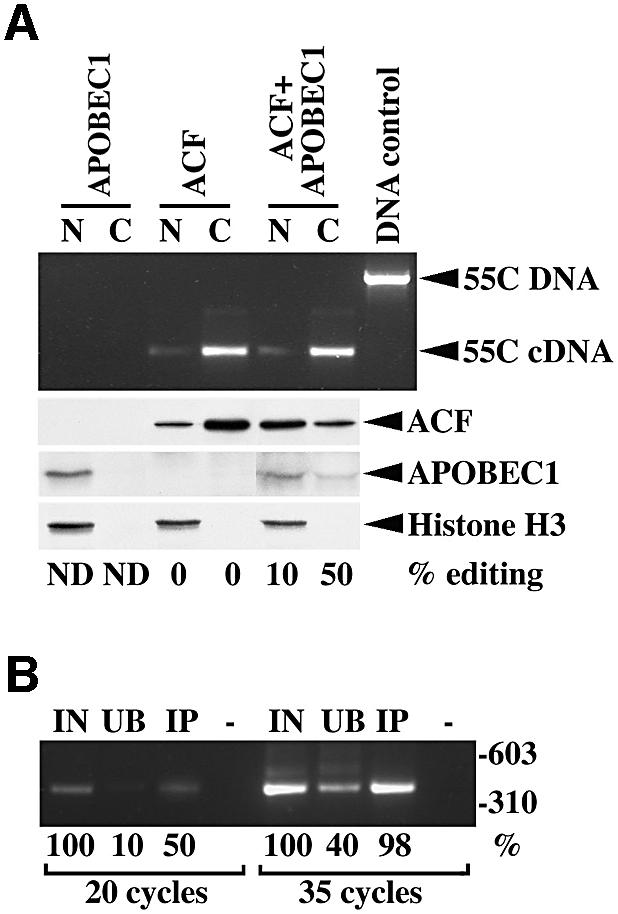
Fig. 6. The APOBEC1–ACF complex contains edited RNA. (A) RT–PCR products (398 bp) derived from immunoprecipitated RNA isolated from nuclear (N) and cytoplasmic (C) extracts obtained by transfecting APOBEC1, ACF or both in CCL13 cells along with the 55C β-globin DNA (55C). Transfected 55C plasmid containing genomic DNA generated a 1390 bp PCR product. Western blot analysis of ACF, APOBEC1 and histone H3 from identical extracts used above and the percentage of apoB RNA editing is shown. (B) RT–PCR (20 and 35 cycles) of input (IN), unbound (UB) and immunoprecipitated (IP) RNA from cytoplasmic extracts containing APOBEC1–ACF. The amount of product is shown as a percentage of the input RNA-derived product for each experimental condition.
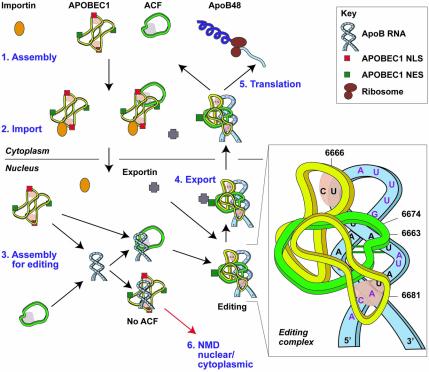
Fig. 7. A model for the assembly, editing and transport of the apoB mRNA editing complex. (1) The initial cytoplasmic event of apoB mRNA editing is the heterotrimerization of ACF with an APOBEC1 dimer. (2) Interaction of importin α with APOBEC1 NLS targets the APOBEC1–ACF complex to the nucleus with ACF as cargo. The importin complex is shown as a single entity consisting of importin α and β subunits. (3) In the nucleus, this complex dissociates or relaxes. Assembly of the editing complex on the apoB RNA occurs via the site-specific binding of ACF to the RNA and the recruitment or repositioning of APOBEC1. C6666 is edited to U, generating a PTC. The editing complex remains associated with the RNA and protects it from NMD. (4) Conformational changes that take place during the assembly of the editing complex expose the APOBEC1 NES. Exportin 1 mediates the export of the apoB RNP complex to the cytoplasm. (5) Translation of edited apoB mRNA generates apoB48. (6) In the absence of ACF, non-specific binding of APOBEC1 to the RNA generates multiple PTCs. RNA with PTC undergoes nuclear or cytoplasmic NMD. The insert shows a schematic representation of the editing complex; unpaired nucleotides are shown in red, APOBEC1 and ACF binding sites are shaded.
Discussion
The results presented here demonstrate that the apoB RNA editing complex serves multiple functions. APOBEC1 transports ACF as cargo to and from the nucleus. ACF binds to the RNA substrate, facilitates the binding of APOBEC1 to the RNA and editing of the substrate C. The association of the APOBEC1–ACF complex with the bona fide edited RNA is essential to protect it from undergoing NMD. APOBEC1 alone generates substrates for NMD. The APOBEC1–ACF complex transports the edited substrate RNA to the cytoplasm for translation.
The evidence presented here strongly supports the view that APOBEC1 is predominantly a nucleocytoplasmic shuttling protein. The APOBEC1 NLS interacts with importin α, and the C-terminal, leucine-rich NES associates with exportin 1 during transport. This finding is consistent with that of Yang et al. (1997, 2001).
ACF apparently does not have an NLS (Blanc et al., 2001b) or an NES (this study), rather it is transported to and from the nucleus by APOBEC1. In the absence of APOBEC1, we found ACF to localize predominantly to the cytoplasm, with some nuclear localization. A similar distribution of ACF was reported recently (Sowden et al., 2002); however, nuclear localization of ACF has also been described (Blanc et al., 2001a). Co-expression of APOBEC1 and ACF localized both proteins to the nucleus. The finding of some ACF in the nucleus suggests that other APOBEC1-independent nuclear import pathways for ACF may also exist. This is consistent with the widespread expression of ACF in tissues that do not express APOBEC1 and its ability to bind RNA, suggesting that ACF may have RNA chaperone activity. Various regions in APOBEC1 and ACF are important for their interaction (Blanc et al., 2001b; Mehta and Driscoll, 2002). We have mapped the interaction of ACF with APOBEC1 to the region of the APOBEC1 NLS, which suggests that the importin α- and ACF-binding sites on APOBEC1 are contiguous or overlapping.
The observation that SV40NLS–APOBEC1(N-30) was unable to enter the nucleus suggested the presence of a potent NES capable of overriding the SV40 NLS. The NES is refined to a 10 amino acid leucine-rich sequence L173–L182 in APOBEC1. Exportin 1 interacts with leucine-rich NESs in nuclear proteins to mediate nuclear export. We demonstrated that APOBEC1 shuttled between the nucleus and cytoplasm in a leptomycin B-dependent manner. We also demonstrated that ACF and APOBEC1 shuttled together, and this was also leptomycin B sensitive. NES mutants of APOBEC1 that failed to shuttle were unable to export ACF. We conclude that APOBEC1 interacts with ACF and transports it both in and out of the nucleus.
We considered whether the APOBEC1–ACF holoenzyme complex might be involved in the export of the edited apoB mRNA from the nucleus. Immuno precipitation of ACF has identified endogenous apoB RNA in the complexes (Mehta et al., 2000; Mehta and Driscoll, 2002), indicating that the complex remains bound to the RNA in vivo. We have extended these observations to identify edited apoB RNA in the nuclear and cytoplasmic complexes. We observed that ∼10% of nuclear RNA and 50% of the cytoplasmic RNA was edited. As expected from our subcellular localization studies, western blot analysis of extracts from cells transfected with APOBEC1 alone identified APOBEC1 in the nucleus, whereas in cells co-transfected with APOBEC1 and ACF, both APOBEC1 and ACF were found mainly in the nucleus and to a lesser extent in the cytoplasm. In cells transfected with ACF alone, ACF was found in both the nucleus and cytoplasm in similar proportions to our immunofluorescence microscopy, which shows ACF to be predominantly cytoplasmic. These findings are supported by the recent ultrastructural analysis of Sowden et al. (2002), which showed that in rodent liver cells expressing endogenous APOBEC1 and ACF, ACF is mainly in the nucleus and to a lesser extent localized to the membranes of the rough endoplasmic reticulum (ER). These results together suggest that the APOBEC1–ACF complex transports the edited apoB mRNA from the nucleus to the ER for translation.
NMD is used to eliminate mRNAs that contain PTCs capable of producing dominant-negative forms of the protein (Hentze and Kulozik, 1999). In mammalian cells, NMD is mainly a cytoplasmic event, but some occurs in the nucleus. The C to U editing of apoB mRNA creates a PTC at codon 2153 in exon 26 (nucleotide C6666). The edited mRNA is stable, does not undergo NMD and produces functional protein.
We have shown that APOBEC1 and ACF in the apoB mRNA editing complex protect the edited RNA from NMD, and transport the RNA to the cytoplasm. APOBEC1 alone does not prevent NMD; rather it produces non-specific hyper-editing, as observed in transgenic mice overexpressing APOBEC1 (Yamanaka et al., 1997), which accelerates NMD. APOBEC1-evoked NMD is protected by the dominant-negative form of hUpf1. This suggests that the NMD mechanism involved utilizes an EJC containing hUpf1, 2 and 3 (Lykke-Andersen et al., 2000). The fact that ACF can protect the edited 55C β-globin mRNA from NMD suggests that ACF can interfere with this process. The observation that PTC 39 and 55T β-globin RNAs are not protected from NMD by ACF suggests that ACF is not directly interfering with the NMD pathway. We hypothesize that the association of the editing complex (APOBEC1–ACF) around the edited site of the RNA masks the PTC generated and may prevent access to the NMD machinery. However, further experiments are necessary to understand the exact role of the editing complex in the protection of the edited apoB mRNA from NMD with respect to EJCs.
The minimal apoB mRNA editing complex, identified to be 120 kDa (Navaratnam et al., 1993a), most probably consists of a single molecule of ACF and an APOBEC1 dimer. We now propose a model in which cytoplasmic ACF interacts with the NLS region on the APOBEC1 monomer through its third RRM (Figure 7). Importin α directs the APOBEC1–ACF complex to the nucleus. In the nucleus, the APOBEC1–ACF complex dissociates or relaxes, and assembly of the editing complex occurs on the apoB RNA. This assembly must utilize the third RRM of ACF and could cause a conformational change in the RNA structure to locate C6666 in a favourable position for editing. The RNA-binding sites of APOBEC1, including the NLS, the active site and the protein–protein interaction domains in both proteins, are involved in the assembly of the editing complex (Figure 7). Subsequently, the RNA is edited and a PTC is generated. Assembly of the editing complex probably exposes the APOBEC1 NES. Interaction of exportin 1 with this NES directs the apoB RNP complex to the cytoplasm. The protein complex protects the apoB RNA with the PTC from undergoing NMD. In the cytoplasm, the RNA is transported to the ER, where a substantial amount of ACF is found (Sowden et al., 2002), and translated to produce apoB48.
In the absence of ACF, the APOBEC1 dimer can bind and edit the RNA at several sites, similar to what is observed in APOBEC1 transgenic animals. This editing can generate multiple PTCs in the RNA. RNA with a PTC can undergo nuclear or cytoplasmic NMD (Figure 7). The functions of the other new members of the APOBEC1 family, including AID, APOBEC2 and APOBEC3A–3G, are unknown. This assay may be used to verify whether these are RNA editing enzymes.
Materials and methods
Plasmids for mammalian expression
APOBEC1 and its mutants with a 3′ FLAG tag were generated by PCR as described (Navaratnam et al., 1995) and cloned into the expression vector pCMV-blue (Pharmingen). ACF and ACFΔ55 with a 3′ c-myc tag were cloned into pcDNA3.1. The β-globin NMD vectors were kindly provided by M.W.Hentze; 55 or 261 nucleotides of human unedited (C) and edited (T) apoB RNA were inserted in-frame into exon 2 of the β-globin wild-type plasmid in the same position as PTC 39.
Cell culture and transfection
COS-7, CCL13, HepG2, McA777 and NIH-3T3 cells (American Type Culture Collection) were maintained as recommended by the supplier. Cells were seeded onto 13 mm round coverslips (9 × 104) or 100 mm dishes (1 × 106) and transfected with 1 µg (coverslips) or up to 55 µg (dishes) of plasmid DNA using the calcium phosphate precipitation method. At 48 h after transfection, the cells were fixed for immunofluorescence microscopy, used for the interspecies heterokaryon assay or harvested for RNA or protein preparation.
Immunofluorescence microscopy
Cells were fixed in 100% methanol for 15 min. Immunostaining was performed with anti-FLAG IgG (Sigma) for APOBEC1, or anti-c-myc IgG (Roche Biochemicals) for ACF, followed by fluorescein isothiocyanate (FITC)-conjugated rabbit anti-mouse IgG (DAKO). For co-localization, the fixed cells were immunostained simultaneously with FITC-conjugated M2 anti-FLAG (Sigma) and Cy3-conjugated c-myc (Sigma), as recommended by the suppliers. The nuclei were stained with 1 µg/ml 4′,6-diamidino-2-phenylindole (DAPI). Slides were visualized under a Leica DMRB fluorescence microscope. The images were captured using IPLab and processed using Adobe PhotoShop 4.0 software. Localization was scored using an unbiased method. A minimum of 100 cells were counted in three independent experiments.
Interspecies heterokaryon assay
Interspecies heterokaryon assays were performed as previously described (Piñol-Roma and Dreyfuss 1992). Briefly, NIH-3T3 cells (1 × 105) were added to transfected human CCL13 cells. After 4 h, the cells were incubated with 50 µg/ml cycloheximide for 15 min. Cell were fused with 50% (w/v) PEG 6000 in phosphate-buffered saline (PBS) for 2–3 min, washed with PBS and incubated at 37°C in the presence of cycloheximide. Where indicated, leptomycin B (50 ng/ml) was added to the co-culture. A 4 h after fusion, heterokaryons were fixed and stained for immunofluorescence microscopy as described above.
Yeast two-hybrid assay
Yeast two-hybrid vectors pJG4-5, pSB202 and pSH18.3-4 were a gift from Roger Brent. Plasmids bearing the importin α, importin β and transportin DNAs, kindly provided by I.Mattaj, and ACF were cloned into pJG4-5. APOBEC1 and mutants were PCR amplified and cloned into pSB202. All methods used in these studies were described previously (Navaratnam et al., 1998).
Northern blot analysis
Northern blot analysis was performed with 8 µg of total RNA extracted from transfected CCL13 as described previously (Jarmuz et al., 2002) and probed with APOBEC1, ACF, hUpf1, lacZ or β-globin cDNA.
RNA immunoprecipitation
Nuclear and cytoplasmic extracts were prepared (Piñol-Roma et al., 1988) from CCL13 cells transiently transfected with 55C together with APOBEC1 and/or ACF. Proteins were immunoprecipitated with anti-FLAG agarose (APOBEC1) or anti-c-myc (ACF) antibody/protein A–agarose. Resins were washed with NET buffer (Mehta and Driscoll, 2002). RNA isolated from the beads with RNAzol (Biogenesis) was amplified by RT–PCR using β-globin gene-specific primers, and TOPO cloned (Invitrogen). Fifty clones were analysed by DNA sequencing. Equivalent protein extracts were also analysed by western blotting with the appropriate antibody. Histone H3 was detected with a monoclonal antibody (Upstate).
Miscellaneous methods
APOBEC1 protein expression and UV cross-linking studies were carried out as described previously (Navaratnam et al., 1993b). In vitro conversion and primer extension analysis of apoB RNA editing were performed as previously described (Navaratnam et al., 1995). Plasmid DNA and oligonucleotide sequences used in this study are available from the authors on request.
Acknowledgments
Acknowledgements
We acknowledge Matthias Hentze and Andreas Kulozik for the β-globin plasmids, and Lynne Maquat for wild-type and mutant human Upf1 plasmids used in this NMD study. We are grateful for to the MRC CSC Photographic and Graphic Design facility for preparing the figures. Work in the RNA Editing Laboratory is supported by the Medical Research Council, UK and Pfizer Inc. (Groton, CT).
References
- Blanc V., Navaratnam,N., Henderson,J.O., Anant,S., Kennedy,S., Jarmuz,A., Scott,J. and Davidson,N.O. (2001a) Identification of GRY-RBP as an apolipoprotein B RNA-binding protein that interacts with both apobec-1 and apobec-1 complementation factor to modulate C to U editing. J. Biol. Chem., 276, 10272–10283. [DOI] [PubMed] [Google Scholar]
- Blanc V., Henderson,J.O., Kennedy,S. and Davidson,N.O. (2001b) Mutagenesis of apobec-1 complementation factor reveals distinct domains that modulate RNA binding, protein–protein interaction with apobec-1 and complementation of C to U RNA-editing activity. J. Biol. Chem., 276, 46386–46393. [DOI] [PubMed] [Google Scholar]
- Chester A., Scott,J., Anant,S. and Navaratnam,N. (2000) RNA editing: cytidine to uridine conversion in apolipoprotein B mRNA. Biochim. Biophys. Acta, 1494, 1–13. [DOI] [PubMed] [Google Scholar]
- Conti E., Uy,M., Leighton,L., Blobel,G. and Kuriyan,J. (1998) Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell, 94, 193–204. [DOI] [PubMed] [Google Scholar]
- Dingwall C. and Laskey,R. (1991) Nuclear targeting sequences—a consensus? Trends Biochem. Sci., 16, 478–481. [DOI] [PubMed] [Google Scholar]
- Fischer U., Huber,J., Boelens,W.C., Mattaj,I.W. and Luhrmann,R. (1995) The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell, 82, 475–483. [DOI] [PubMed] [Google Scholar]
- Flach J., Bossie,M., Vogel,J., Corbett,A., Jinks,T., Willins,D.A. and Silver,P.A. (1994) A yeast RNA binding protein shuttles between the nucleus and the cytoplasm. Mol. Cell. Biol., 14, 8399–8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M., Ohno,M., Yoshida,M. and Mattaj,I.W. (1997) CRM1 is an export receptor for leucine-rich nuclear export signals. Cell, 90, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Fridell R.A., Truant,R., Thorne,L., Benson,R.E. and Cullen,B.R. (1997) Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-β. J. Cell Sci., 110, 1325–1331. [DOI] [PubMed] [Google Scholar]
- Frischmeyer P.A. and Dietz,H.C. (1999) Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet., 8, 1893–1900. [DOI] [PubMed] [Google Scholar]
- Fujino T., Navaratnam,N., Jarmuz,A., von Haeseler,A. and Scott,J. (1999) C–U editing of apolipoprotein B mRNA in marsupials: identification and characterisation of APOBEC-1 from the American opossum Monodelphus domestica. Nucleic Acids Res., 27, 2662–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A.P. and Keller,W. (2001) RNA editing by base deamination: more enzymes, more targets, new mysteries. Trends Biochem. Sci., 26, 376–384. [DOI] [PubMed] [Google Scholar]
- Görlich D. and Mattaj,I.W. (1996) Nucleocytoplasmic transport. Science, 271, 1513–1518. [DOI] [PubMed] [Google Scholar]
- Hentze M.W. (2001) Protein synthesis. Believe it or not—translation in the nucleus. Science, 293, 1058–1059. [DOI] [PubMed] [Google Scholar]
- Hentze M.W. and Kulozik,A.E. (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell, 96, 307–310. [DOI] [PubMed] [Google Scholar]
- Hersberger M. and Innerarity,T.L. (1998) Two efficiency elements flanking the editing site of cytidine 6666 in the apolipoprotein B mRNA support mooring-dependent editing. J. Biol. Chem., 273, 9435–9442. [DOI] [PubMed] [Google Scholar]
- Iborra F.J., Jackson,D.A. and Cook,P.R. (2001) Coupled transcription and translation within nuclei of mammalian cells. Science, 293, 1139–1142. [DOI] [PubMed] [Google Scholar]
- Jarmuz A., Chester,A., Bayliss,J., Gisbourne,J., Dunham,I., Scott,J. and Navaratnam,N. (2002) An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics, 79, 285–296. [DOI] [PubMed] [Google Scholar]
- La Casse E.C. and Lefebvre,Y.A. (1995) Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res., 23, 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P.P., Xiong,W.J., Zhu,H.J., Chen,S.H. and Chan,L. (1991) Apolipoprotein B mRNA editing is an intranuclear event that occurs posttranscriptionally coincident with splicing and polyadenylation. J. Biol. Chem., 266, 20550–20554. [PubMed] [Google Scholar]
- Le Hir H., Izaurralde,E., Maquat,L.E. and Moore,M.J. (2000) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J., 19, 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lellek H., Kirsten,R., Diehl,I., Apostel,F., Buck,F. and Greeve,J. (2000) Purification and molecular cloning of a novel essential component of the apolipoprotein B mRNA editing enzyme-complex. J. Biol. Chem., 275, 19848–19856. [DOI] [PubMed] [Google Scholar]
- Liao W., Hong,S.-H., Chan,B.H.-J., Rudolph,F.B., Clark,S.C. and Chan,L. (1999) APOBEC-2, a cardiac- and skeletal muscle-specific member of the cytidine deaminase supergene family. Biochem. Biophys. Res. Commun., 260, 398–404. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu,M.D. and Steitz,J.A. (2000) Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell, 103, 1121–1131. [DOI] [PubMed] [Google Scholar]
- Madsen P. et al. (1999) Psoriasis upregulated phorbolin-1 shares structural but not functional similarity to the mRNA-editing protein Apobec-1. J. Invest. Dermatol., 113, 162–169. [DOI] [PubMed] [Google Scholar]
- Maquat L.E. (1996) Defects in RNA splicing and the consequence of shortened translational reading frames. Am. J. Hum. Genet., 59, 279–286. [PMC free article] [PubMed] [Google Scholar]
- Maquat L.E. and Carmichael,G.G. (2001) Quality control of mRNA function. Cell, 104, 173–176. [DOI] [PubMed] [Google Scholar]
- Mehta A. and Driscoll,D.M. (2002) Identification of domains in apobec-1 complementation factor required for RNA binding and apolipoprotein B mRNA editing. RNA, 8, 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A., Kinter,M.T., Sherman,N.E. and Driscoll,D.M. (2000) Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol. Cell. Biol., 20, 1846–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M., Sankaranand,V.S., Anant,S., Susgai,M., Kinoshita,K., Davidson,N.O. and Honjo,T. (1999) Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem., 274, 18470–18476. [DOI] [PubMed] [Google Scholar]
- Nagy E. and Maquat,L.E. (1998) A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci., 23, 198–199. [DOI] [PubMed] [Google Scholar]
- Navaratnam N., Shah,R., Patel,D., Fay,V. and Scott,J. (1993a) Apolipoprotein B mRNA editing is associated with UV crosslinking of proteins to the editing site. Proc. Natl Acad. Sci. USA, 90, 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaratnam N., Morrison,J.R., Bhattycharya,S., Patel,D., Funahashi,T., Giannoni,F., Teng,B.-B., Davidson,N.O. and Scott,J. (1993b) The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J. Biol. Chem., 268, 20709–20712. [PubMed] [Google Scholar]
- Navaratnam N., Bhattycharya,S., Fujino,T., Patel,D., Jarmuz,A.L. and Scott,J. (1995) Evolutionary origins of apoB mRNA editing: catalysis by a cytidine deaminase that has acquired a novel RNA-binding motif at its active site. Cell, 81, 187–195. [DOI] [PubMed] [Google Scholar]
- Navaratnam N. et al. (1998) Escherichia coli cytidine deaminase provides a molecular model for apoB RNA editing and a mechanism for RNA substrate recognition. J. Mol. Biol., 275, 695–714. [DOI] [PubMed] [Google Scholar]
- Piñol-Roma S. and Dreyfuss,G. (1992) Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature, 355, 730–732. [DOI] [PubMed] [Google Scholar]
- Piñol-Roma S., Choi,Y.D., Matunis,M.J. and Dreyfuss,G. (1988) Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev., 2, 215–227. [DOI] [PubMed] [Google Scholar]
- Pollard V.W. and Malim,M.H. (1998) The HIV-1 Rev protein. Annu. Rev. Microbiol., 52, 491–532. [DOI] [PubMed] [Google Scholar]
- Richardson N., Navaratnam,N. and Scott,J. (1998) Secondary structure for the apolipoprotein B mRNA editing site: AU-binding proteins interact with a stem loop. J. Biol. Chem., 273, 31707–31717. [DOI] [PubMed] [Google Scholar]
- Schell T., Kulozik,A.E. and Hentze,M.W. (2002) Integration of splicing, transport and translation to achieve mRNA quality control by the nonsense-mediated decay pathway. Genome Biol., 3, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. (1995) A place in the world for RNA editing. Cell, 81, 833–836. [DOI] [PubMed] [Google Scholar]
- Shah R.R., Knott,T.J., LeGros,J.E., Navaratnam,N., Greeve,J.C. and Scott,J. (1991) Sequence requirements for the editing of apolipoprotein B mRNA. J. Biol. Chem., 266, 16301–16304. [PubMed] [Google Scholar]
- Sheehy A.M., Gaddis,N.C., Choi,J.D. and Malim,M.H. (2002) Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature, 418, 646–650. [DOI] [PubMed] [Google Scholar]
- Sowden M.P., Ballatori,N., de Mesy Jensen,K.L., Hamilton Reed,L. and Smith,H.C. (2002) The editosome for cytidine to uridine mRNA editing has a native complexity of 27S: identification of intracellular domains containing active and inactive editing factors. J. Cell Sci. 115, 1027–1039. [DOI] [PubMed] [Google Scholar]
- Sun X., Perlick,H.A., Dietz,H.C. and Maquat,L.M. (1998) A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl Acad. Sci. USA, 95, 10009–10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng B.-B., Burant,C.F. and Davidson,N.O. (1993) Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science, 260, 1816–1819. [DOI] [PubMed] [Google Scholar]
- Thermann R., Neu-Yilik,G., Deters,A., Frede,U., Wehr,K., Hagemeier,C., Hentze,M.W. and Kulozik,A.E. (1998) Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J., 17, 3484–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff B., Sanglier,J.-J. and Wang,Y. (1997) Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol., 4, 139–147. [DOI] [PubMed] [Google Scholar]
- Yamanaka S., Poksay,K.S., Arnold,K.S. and Innerarity,T.L. (1997) A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes Dev., 11, 321–333. [DOI] [PubMed] [Google Scholar]
- Yang Y., Yang,Y. and Smith,H.C. (1997) Multiple protein domains determine the cell type-specific nuclear distribution of the catalytic subunit required for apolipoprotein B mRNA editing. Proc. Natl Acad. Sci. USA, 94, 13075–13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Sowden,M.P., Yang,Y. and Smith,H.C. (2001) Intracellular trafficking determinants in APOBEC-1, the catalytic subunit for cytidine to uridine editing of apolipoprotein B mRNA. Exp. Cell Res., 267, 153–164. [DOI] [PubMed] [Google Scholar]