Abstract
The activation of the transcription factor NF-κB is central to the control of the cellular response triggered by many stimuli. Once released from the inhibitory molecule IκB, NF-κB is translocated to the nucleus, and it has to be phosphorylated to activate transcription. In ζ protein kinase C (PKC)-deficient cells, NF-κB is transcriptionally inactive and the phosphorylation of the RelA subunit in response to tumor necrosis factor (TNF-α) is severely impaired. In vitro assays showed that ζPKC directly phosphorylates RelA. Here we demonstrate that Ser311 accounts for ζPKC phosphorylation of RelA and that this site is phosphorylated in vivo in response to TNF-α. Also, an inactivating mutation of that residue severely impairs RelA transcriptional activity, blocks its anti-apoptotic function and abrogates the interaction of RelA with the co-activator CBP as well as its recruitment, and that of RNA polymerase II (Pol II) with the interleukin-6 (IL-6) promoter. The interaction of endogenous CBP with endogenous RelA is inhibited in ζPKC–/– cells, as well as the binding of Pol II to the IL-6 promoter. These results demonstrate the mechanism whereby ζPKC regulates NF-κB activation in vivo.
Keywords: NF-κB/phosphorylation/ζPKC/RelA/transcription activation
Introduction
Nuclear factor κB (NF-κB) complexes are essential for the control of a large variety of cellular functions including the synthesis of inflammatory cytokines and cell survival, both central to the control of the immune response and in tumor transformation (Karin et al., 2002; Li and Verma, 2002; Moscat et al., 2003). NF-κBs are dimers of various combinations of Rel proteins which include RelA (p65), c-Rel, RelB, NF-κB1 (p105) and NF-κB2 (p100) (Ghosh and Karin, 2002). The proteolytic processing of NF-κB1 and NF-κB2 generates p50 and p52, respectively. The complex p65/p50 is retained in the cytoplasm by the inhibitory protein IκB whose phosphorylation by the IκB kinase (IKK) β component of the IKK signalsome complex leads to its ubiquitylation and proteasome-mediated degradation (Ghosh and Karin, 2002). This produces the release of the p65/p50 dimer that enters the nucleus, triggering the transcription of a large number of genes. This pathway is activated by inflammatory cytokines such as tumor necrosis factor-α (TNF-α). Parallel to this classical cascade, a novel mechanism to generate a different NF-κB complex has been uncovered recently. This is composed of the transactivating subunit RelB which is bound to p100 and kept inactive in the cytosol. Upon stimulation of fibroblasts with lymphotoxin-β, or B cells with BAFF, the IKKα subunit of the IKK complex phosphorylates p100, triggering its processing and the release of the RelB–p52 complex that now is free to enter the nucleus (Senftleben et al., 2001; Xiao et al., 2001; Claudio et al., 2002; Dejardin et al., 2002; Kayagaki et al., 2002).
A number of reported pieces of evidence indicates that once in the nucleus, RelA has to be phosphorylated to be transcriptionally competent. In this regard, Ghosh’s laboratory identified protein kinase A (PKA) as a RelA-associated kinase that was able to phosphorylate Ser276 in the Rel homology domain (RHD) of RelA once IκB is degraded and RelA is made accessible to the kinase (Zhong et al., 1997). This phosphorylation event has been shown to be important for RelA transcriptional activity (Zhong et al., 1997). Two other sites have been proposed to be phosphorylated in RelA. Both are located not in the RHD but in the transactivation domain. Thus, the phosphorylation of Ser529 was claimed to be necessary for RelA transcriptional activity in Ras-transformed, as well as in TNF-α-activated cells (Wang and Baldwin, 1998). Recently, casein kinase II (CKII) has been proposed as the kinase responsible for Ser529 phosphorylation which, like PKA’s actions on Ser276, requires prior release of p65/p50 from the IκB complex (Wang et al., 2000). Another potentially phosphorylated site located in the RelA transactivation domain is Ser536 (Sakurai et al., 1999). The transactivation domain of RelA appears to be targeted by phosphoinositide 3-kinase-derived signals (Sizemore et al., 1999; Madrid et al., 2000), and the phosphorylation of Ser536 has been reported to be mediated by IKKβ in some systems (Sakurai et al., 1999). GSK-3β and T2K are two kinases which have also been implicated in the control of NF-κB transcriptional activity, but it is still unclear whether RelA phosphorylation is a target of their mechanism of action (Bonnard et al., 2000; Hoeflich et al., 2000).
The two members of the atypical subfamily of protein kinase C (aPKC), namely ζPKC and λ/ιPKC, have been implicated in co-transfection experiments in the activation of NF-κB in response to several stimuli (Lallena et al., 1999; Moscat and Diaz-Meco, 2000). We recently have characterized ζPKC knockout mice and have established that this kinase is actually important for the activation of NF-κB in vivo. Although in co-transfection assays ζPKC is able to modulate IKK activity, in ζPKC–/– mice it only appears to be essential for IKK activation in the lung in in vivo experiments, an organ in which ζPKC is particularly abundant (Leitges et al., 2001). In B cells, it is clear that the loss of ζPKC inhibits the synthesis of a number of κB-dependent genes with no effects on NF-κB nuclear translocation, as determined by electrophoretic mobility shift assays (EMSAs) (Martin et al., 2002). In embryo fibroblasts (EFs), the loss of ζPKC also impairs the synthesis of κB-dependent genes but not the translocation of NF-κB or the activation of the IKK complex (Leitges et al., 2001). More importantly, we showed that phosphorylation of RelA in response to TNF-α was severely inhibited in ζPKC–/– EFs, strongly suggesting that ζPKC directly controls RelA phosphorylation. In fact, we showed that ζPKC is capable of directly phosphorylating the RHD of RelA in vitro (Leitges et al., 2001). In the present study, we have mapped the ζPKC phosphorylation site and have determined that it actually is a bona fide endogenous site whose phosphorylation is required for RelA function.
Results
Identification of the ζPKC phosphorylation site in vitro
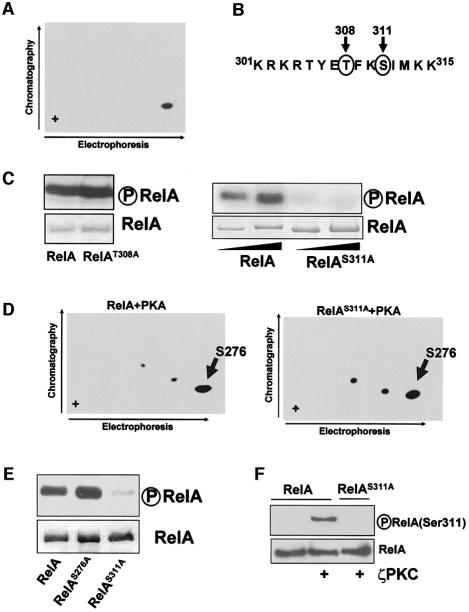
Since ζPKC has been shown to contribute to RelA phosphorylation in vivo and to phosphorylate the RHD of RelA directly in vitro, we initially determined the number of peptides phosphorylated by recombinant ζPKC in an in vitro phosphorylation assay using bacterially expressed recombinant pure RelA RHD, followed by tryptic peptide mapping. Note that the average stoichiometry of this phosphorylation is 0.8 ± 0.1 mol phosphate/mol protein. Phosphopeptides from phosphorylated RelA were analyzed by two-dimensional separation on thin layer cellulose plates, which gave a pattern consistent with ζPKC phosphorylating a single peptide in RelA (Figure 1A). In order to identify the site(s) targeted by ζPKC in this assay, we searched the RelA sequence with the protein database motif engine Scansite (http://scansite.mit.edu) to identify putative ζPKC phosphorylation sites in RelA. Two potential residues, Thr308 and Ser311, gave a significant score (Figure 1B). It should be noted that both sites were conserved in human and murine RelA. In order to determine which of these sites could account for RelA phosphorylation by ζPKC, both residues were mutated individually to alanine, and mutant recombinant RHD RelA proteins were bacterially expressed and tested in an in vitro phosphorylation assay with recombinant ζPKC. The results in Figure 1C demonstrate that mutation of Thr308 did not affect RelA phosphorylation by ζPKC. In contrast, mutation of Ser311 completely abrogated RelA phosphorylation by ζPKC (Figure 1D). Ser276 has been reported to be phosphorylated by PKA in vitro and in vivo (Zhong et al., 1997). To demonstrate that the S311A mutant does not affect the phosphorylation of Ser276, either wild-type or S311A mutant RelA RHD was phosphorylated by PKA in vitro, after which the tryptic phosphopeptides were analyzed by two-dimensional separation as above. The results in Figure 1D (left panel) show that PKA phosphorylates one main and two minor peptides. Comparison of this map with that of the RHD S276A mutant (not shown) reveals that the major peptide contains Ser276. It is of note that none of these two minor sites coincide with the ζPKC phosphorylated peptide (not shown). Interestingly, the ability of PKA to phosphorylate Ser276 is not affected by the mutation of Ser311 (Figure 1D, right panel). Similarly, the S276A mutation does not impair RelA phosphorylation by ζPKC (Figure 1E).
Fig. 1. Mapping of the ζPKC phosphorylation site in RelA. (A) Recombinant RelA RHD was phosphorylated in vitro with recombinant baculovirus ζPKC. The RelA tryptic phosphopeptides were separated by electrophoresis followed by ascending chromatography. (B) Putative ζPKC phosphorylation sites in RelA as predicted by the Scansite program. (C) Recombinant RelA RHD either wild-type (RelA, left and right panels) or mutated (RelAT308A, left panel; RelAS311A, right panel) was phosphorylated in vitro by recombinant ζPKC. (D) Recombinant RelA RHD either wild-type or S311A mutant was phosphorylated in vitro with pure PKA and the tryptic peptides were separated by electrophoresis followed by ascending chromatography as above. (E) Recombinant RelA RHD either wild-type or mutated at Ser311 or Ser276 was phosphorylated in vitro with ζPKC as above. (F) Recombinant RelA wild-type or S311A mutant was incubated in phosphorylation reactions with ζPKC, after which the phosphorylation of Ser311 was determined by immunoblotting with the anti-phospho-RelA(Ser311) antibody. As a negative control, wild-type RelA was incubated in the absence of ζPKC.
Ser311 is a bona fide endogenous phosphorylation site
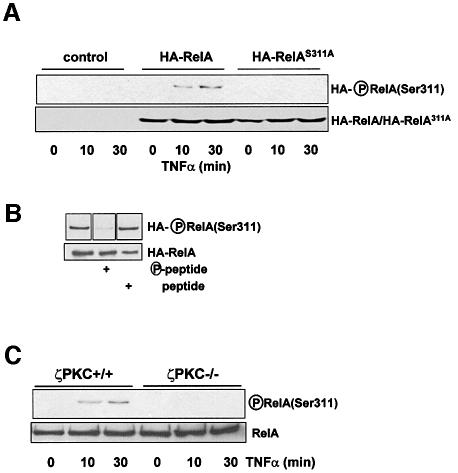
Once Ser311 has been shown to be the in vitro ζPKC phosphorylation site in RelA, we sought to determine whether this residue is actually phosphorylated in vivo. To address this question, we prepared an anti-phospho-RelA(Ser311) antibody. We first confirmed that this reagent is in fact specific for this site. Thus, recombinant wild-type RelA or the RelAS311A mutant were phosphorylated in vitro by ζPKC, after which reaction mixtures were analyzed by immunoblotting with the anti-phospho-RelA(Ser311) antibody. This antibody reacted with the wild-type protein phosphorylated by ζPKC but not with the unphosphorylated protein or with the S311A mutant that has been incubated with ζPKC (Figure 1F). We next stably expressed either hemagglutinin (HA)-tagged wild-type RelA or the RelAS311A mutant form in EFs by using a pBabe-based retroviral system. These cells were stimulated with TNF-α for different times, after which cells extracts were immunoprecipitated with an anti-HA antibody and the immunoprecipitates were analyzed by immunoblotting with the anti-phospho-RelA(Ser311) antibody. The data in Figure 2A (upper panel) demonstrate that ectopically expressed wild-type RelA was phosphorylated at Ser311, whereas the RelAS311A mutant was not. The lower panel of Figure 2A shows that the levels of wild-type and mutant RelA were comparable in this experiment. The experiment in Figure 2B shows that incubation with the phospho-peptide used to generate the antibody, but not with the dephospho-peptide, inhibited the reaction of the antibody with the RelA phosphorylated protein. In order to investigate whether this site is actually phosphorylated in endogenous RelA, and that this is dependent on ζPKC, either wild-type or ζPKC knockout EFs were stimulated with TNF-α for different times, after which endogenous RelA was immunoprecipitated and the phosphorylation of Ser311 was determined in the immunoprecipitates by immunoblotting with the anti-phospho-RelA(Ser311) antibody. Of potential great relevance, endogenous RelA is phosphorylated at Ser311 in a TNF-α-dependent manner in wild-type EFs and, more importantly, this phosphorylation is severely abrogated in ζPKC–/– EFs (Figure 2C).
Fig. 2. Phosphorylation of RelA Ser311 in vivo. (A) Control EFs or stably expressing HA-tagged wild-type or S311A mutant RelA were incubated with TNF-α for different times, after which the ectopically expressed RelA was immunoprecipitated with the anti-HA antibody and Ser311 phosphorylation was determined by immunoblotting with the anti-phospho-RelA(Ser311) antibody (upper panel). The lower panel shows a parallel immunoblot with the anti-HA antibody. (B) The immunoprecipitates from the cells that had been treated with TNF-α for 30 min were immunoblotted with the anti-phospho-RelA(Ser311) antibody in the absence or presence of phospho- or dephospho-peptide. (C) Wild-type or ζPKC-deficient EFs were stimulated with TNF-α for different times, after which endogenous RelA was immunoprecipitated and Ser311 phosphorylation was determined by immunoblotting as above.
Functional role of Ser311 phosphorylation for NF-κB transcriptional activity
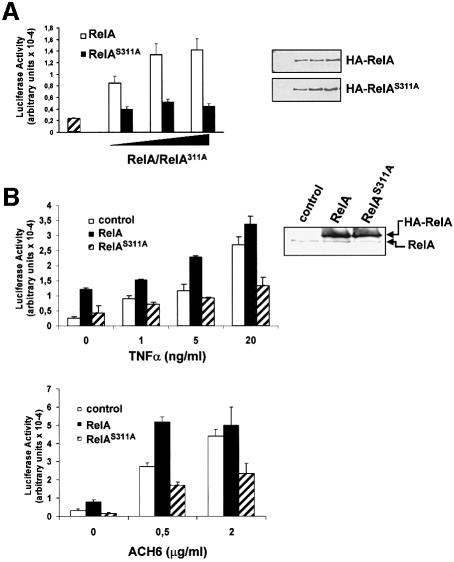
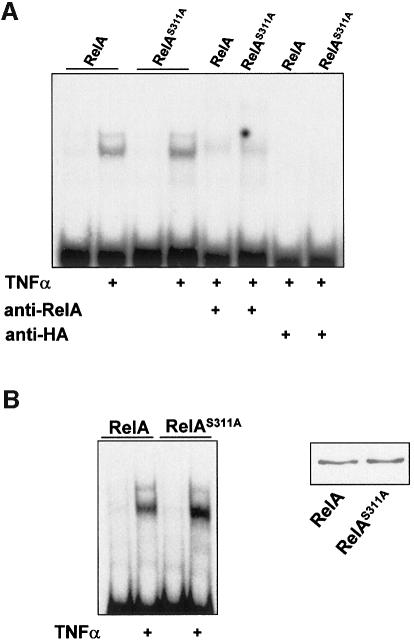
In order to determine the potential functional relevance of Ser311 phosphorylation, increasing amounts of HA-tagged full-length RelA, either wild-type or mutated (RelAS311A), were transfected into 293 cells along with a κB luciferase reporter plasmid and a Renilla control reporter, to normalize for transfection efficiencies. Interestingly, although the expression of increasing amounts of wild-type RelA produces, as expected, the activation of NF-κB-dependent transcription (Figure 3A), the expression of comparable amounts of RelAS311A was unable to activate the κB-dependent reporter activity. Qualitatively similar results were obtained in a similar experiment performed in TNF-α-treated cells (not shown). This indicates that phosphorylation of Ser311 is functionally important. In order to confirm this conclusion further, the cell lines that stably overexpress the HA-tagged versions of wild-type and RelAS311A were transfected with the κB-luciferase reporter along with the Renilla plasmid control and were stimulated with different concentrations of TNF-α (Figure 3B), interleukin (IL)-1 (not shown) or the anti-lymphotoxin-β receptor agonistic antibody ACH6 (Figure 3B), after which κB-dependent transcription was determined. It is of note that cells that were transduced with the empty pBabe control vector responded to agonist-induced κB-dependent transcription (Figure 3B). Cells that ectopically expressed wild-type RelA responded better than the controls, particularly at low agonist concentrations (Figure 3B). Interestingly, cells that stably express the RelAS311A mutant showed a dramatically inhibited response to the different tested agonists, indicating that phosphorylation of Ser311 is essential for κB-dependent transcription. The results in Figure 3B, upper right panel, demonstrate that the expression levels of the mutant RelA were comparable with those of the wild-type protein. Despite the impairment in κB-dependent transcription, NF-κB nuclear translocation and interaction with the κB oligonucleotide probe in the RelAS311A cells were not affected. Thus, EMSAs were performed in the TNF-α-activated wild-type and S311A stably transduced cell lines. The results in Figure 4A demonstrate that both anti-RelA and anti-HA antibodies were capable of supershifting/inhibiting the NF-κB complex, indicating that most of the NF-κB signal in these cells is accounted for by the ectopically expressed protein. When RelA and RelAS311A were introduced in RelA–/– cells and these were activated with TNF-α, it become clear that both the wild-type and the mutant RelA protein were capable of interacting with the κB oligonucleotide probe (Figure 4B), indicating that the defects in κB-dependent transcription observed in the RelA mutant-expressing cells cannot be explained by hypothetical alterations of DNA binding. It should be noted that also in the background of the RelA knockout cells, RelAS311A was unable to activate transcription properly compared with the wild-type protein (not shown). However, AP1 transcriptional activity or that of a cytomegalovirus (CMV)-based constitutive promoter was not affected by expression of the mutant RelA (not shown). Our previous data demonstrated that the loss of ζPKC dramatically inhibited the synthesis of IL-6 RNA in response to TNF-α (Leitges et al., 2001). The data in Figure 5 show that IL-6 production as determined by enzyme-linked immunosorbent assay (ELISA) was impaired not only in the ζPKC–/– EFs (Figure 5A) but also in EFs stably expressing the RelAS311A mutant, compared with their respective controls (Figure 5B). It is of note that cell activation with forskolin, which should target Ser276 through PKA, is not affected in the S311A mutant cell line (see the Supplementary data available at The EMBO Journal Online). Similarly, the activation of NF-κB transcription by overexpression of ζPKC is not inhibited by expression of the S276A mutant (not shown), indicating that both residues may act independently.
Fig. 3. Role of Ser311 in NF-κB transcription. (A) Subconfluent cultures of 293 cells were transfected with either empty plasmid (striped bar) or different amounts of expression vectors for HA-tagged wild-type or S311A mutant RelA along with a κB-dependent reporter and the Renilla control plasmid. Luciferase activity was determined as described in Materials and methods. Results are the mean ± SD of three independent experiments with incubations in duplicate (left panel). The expression levels of wild-type and mutant RelA of one representative experiment were determined by immunoblotting with an anti-HA antibody (right panel). (B) EFs transduced with empty pBabe vector (control) or stably expressing wild-type or S311A mutant RelA were transfected with the κB-luciferase reporter vector along with the control Renilla plasmid, after which they were stimulated for 6 h with different concentrations of TNF-α (upper left panel) or the agonistic anti-lymphotoxin-β receptor antibody ACH6 (lower panel). Luciferase activity was determined as above. Expression levels of endogenous and ectopically expressed RelA proteins were determined by immunoblotting with an anti-RelA antibody (upper right panel).
Fig. 4. Lack of effect of S311A mutation on NF-κB nuclear activity. (A) Nuclear extracts from cells expressing wild-type or the S311A mutant that had been stimulated or not with TNF-α for 1 h were analyzed by EMSA using a κB probe. These nuclear extracts were also pre-incubated with anti-RelA or anti-HA antibodies to determine the contribution of the ectopically expressed RelA proteins to the NF-κB complex. (B) RelA–/– cells that had been transduced with wild-type or mutant RelA proteins were stimulated or not with TNF-α for 1 h and nuclear extracts analyzed by EMSA as above (upper panel), or total cell extracts were analyzed by immunoblotting with anti-HA antibody.
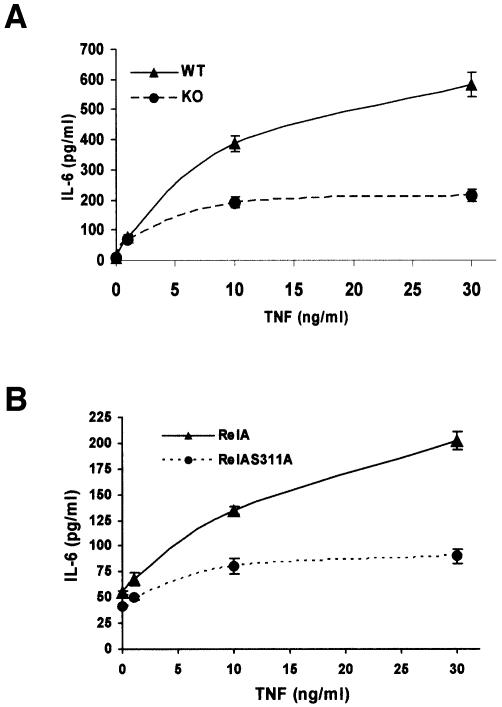
Fig. 5. Control of IL-6 synthesis by ζPKC and RelA Ser311 phosphorylation. (A) EFs from wild-type (WT) or ζPKC-deficient mice (KO) were stimulated with different concentrations of TNF-α for 24 h, after which IL-6 production was determined by ELISA. (B) IL-6 production was also determined in EFs stably expressing WT or the S311A RelA mutant. Results are the mean ± SD of two independent experiments with incubations in duplicate.
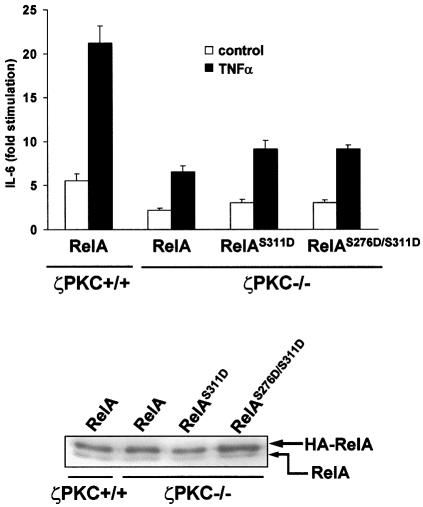
Mutation of serine to aspartate creates a negative charge in the residue that in part may mimic the environment of the phosphorylated serine. In order to determine whether a S311D mutation would rescue RelA transcriptional activity in ζPKC–/– cells, either wild-type or ζPKC-deficient EFs were transfected with wild-type RelA or the mutants RelAS311D or RelAS276D/S311D. Cells were stimulated or not with TNF-α, and IL-6 production was determined as above. The ability of RelA to trigger IL-6 synthesis was clearly blunted in the ζPKC-deficient EFs compared with the wild-type controls (Figure 6). Mutation of Ser311 to aspartate, either alone or in combination with the S276D mutant, was unable to rescue the loss of ζPKC (Figure 6). These results suggest that either the serine to aspartate mutation cannot completely mimic Ser311 and Ser276 phosphorylation, or that ζPKC actions other than Ser311 phosphorylation are required for a full RelA transcriptional activity. In this regard, it should be noted that acidic substitutions of serines as a way to mimic their phosphorylation state may be limited. Thus, for example, when the two activating serines of the kinase MEK were mutated to acidic residues, the mutated MEK only displayed 0.5% of the activity of the actually phosphorylated enzyme (Cowley et al., 1994). Therefore, it is possible that the very partial changes induced by the S311D mutation may not be sufficient to mimic the conformational alterations produced by the phosphorylation of Ser311 required for its transcriptional activity.
Fig. 6. ζPKC is required for RelA-induced IL-6 synthesis. Either wild-type or ζPKC-deficient EFs were transfected with wild-type RelA or the different mutants, after which cells were stimulated or not with TNF-α, and IL-6 synthesis was determined as above (upper panel). Immunoblot analysis with anti-RelA antibody shows the expression levels of the transfected and endogenous RelA protein.
Mutation of Ser311 inhibits the pro-survival role of RelA
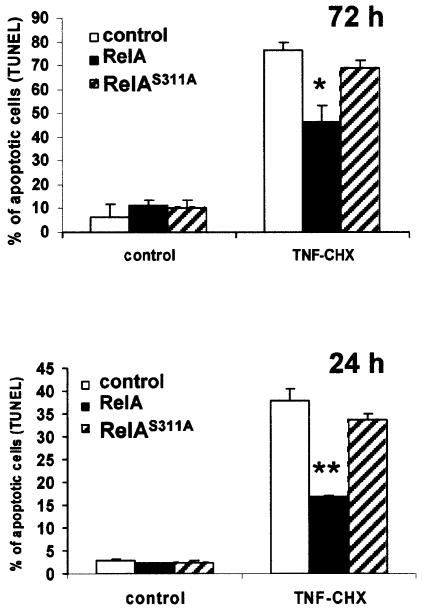
TNF-α is a potent inducer of apoptosis as long as NF-κB production is inhibited (Karin and Lin, 2002). In order to determine whether the loss of function produced by Ser311 mutation to activate κB-dependent transcription is also reflected in the ability of TNF-α to induce apoptosis, pBabe-transduced cells were treated or not with TNF-α plus a limiting amount of cycloheximide that is sufficient to allow TNF-α to induce apoptosis but does not completely block protein synthesis (Tang et al., 2001). Interestingly, control cells underwent apoptosis at 24 and 72 h in response to TNF-α very efficiently, as expected (Figure 7). However, cells stably overexpressing wild-type RelA were much less efficient than control cells in undergoing cell death, consistent with the notion that NF-κB is an anti-apoptotic transcription factor (Figure 7). Importantly, TNF-α-induced apoptosis of cells expressing RelAS311A was comparable with that of the control cells (Figure 7), indicating that mutation of Ser311 leads to the loss of the ability of overexpressed RelA to restrain cell death in response to activation of the TNF-α pathway. The differences between the wild-type and mutant RelA were more apparent when apoptosis was determined at 24 h than at 72 h, although the differences were statistically significant at both times (Figure 7). Collectively, these results suggest that phosphorylation of Ser311 is crucial for RelA to activate NF-κB-dependent transcription and to prevent apoptosis, a hallmark of NF-κB function.
Fig. 7. Role of Ser311 in NF-κB-induced survival. Control EFs or stably expressing wild-type or S311A RelA mutant were incubated in the absence or presence of TNF-α plus cycloheximide for 72 (upper panel) or 24 h (lower panel), after which the percentage of cells undergoing apoptosis was determined by TUNEL analysis. Results are the mean ± SD of two independent experiments with incubations in duplicate. P-values were calculated according to Student’s t-test. *P < 0.01; **P < 0.001.
Impaired RelA–CBP functional and physical interaction by S311A mutation in vivo
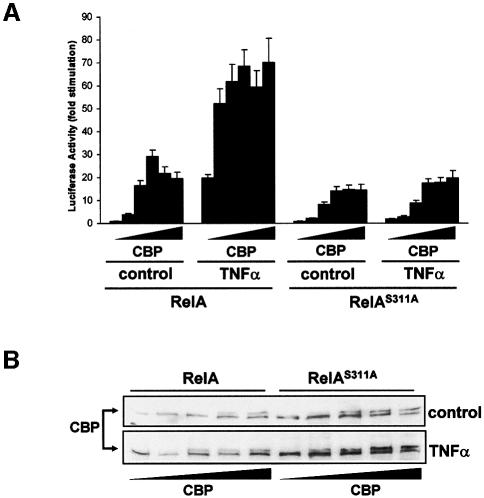
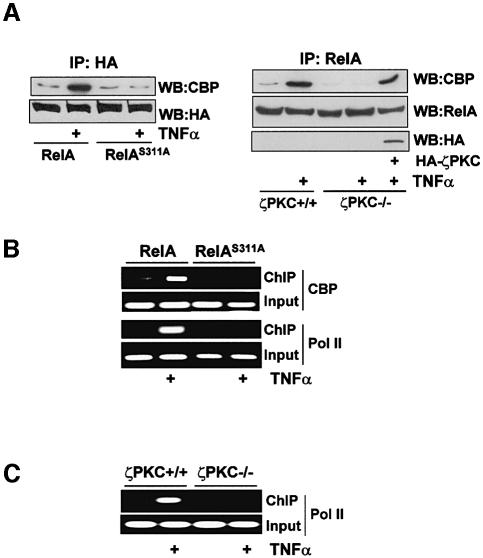
The interaction of phosphorylated RelA with the co-activator CBP/p300 is essential for κB-dependent transcription (Zhong et al., 1998). As the mutation of Ser311 impairs its transcriptional activity, we sought to determine whether the ability of RelA to cooperate with transfected CBP to activate κB-dependent transcription was affected in the RelAS311A mutant. The results in Figure 8A demonstrate that increasing concentrations of CBP significantly enhanced basal and TNF-α-induced NF-κB transcriptional activity in cells that stably express wild-type RelA. However, when CBP was transfected into cells stably expressing the RelAS311A mutant, its ability to cooperate in the activation of κB-dependent transcription was severely impaired, indicating that phosphorylation of Ser311 is required for RelA to interact functionally with CBP. The results in Figure 8B demonstrate that the inability of CBP to activate the NF-κB transcriptional activity in the RelAS311A mutant cells is not due to lower expression. In order to establish whether this mutation affected the physical interaction of RelA with endogenous CBP, cells stably expressing wild-type RelA or the S311A mutant were stimulated or not with TNF-α, after which cell extracts were immunoprecipitated with the anti-HA antibody and the immunoprecipitates were analyzed by immunoblotting with an anti-CBP antibody. Interestingly, stimulation with TNF-α promoted the interaction of wild-type RelA with endogenous CBP (Figure 9A, left panel), in agreement with previously reported data (Zhong et al., 1998). However, the interaction of endogenous CBP with RelAS311A was dramatically reduced compared with that of wild-type RelA (Figure 9A, left panel). Importantly, the interaction of endogenous CBP with endogenous RelA in TNF-α-activated cells is inhibited by the loss of ζPKC (Figure 9A, right panel) but is rescued when HA-ζPKC was ectopically reintroduced into the ζPKC–/– cells. Therefore, the interaction of RelA with CBP requires the phosphorylation of Ser311 by ζPKC. We next performed chromatin immunoprecipitation (ChIP) experiments to determine whether the S311A mutation actually weakens the recruitment of CBP to the endogenous IL-6 promoter in vivo. The data in Figure 9B clearly show that TNF-α stimulation of cells expressing wild-type RelA provokes a reproducible recruitment of endogenous CBP to the IL-6 promoter. Interestingly, this recruitment is severely abrogated in the RelAS311A cells (Figure 9B). In order to determine whether the impairment of CBP binding to the IL-6 promoter detected in the RelAS311A-expressing cells inhibits the recruitment of RNA polymerase II (Pol II), ChIP experiments were performed in these cells using an anti-Pol II antibody. It is clear from the data in Figure 9B that Pol II recruitment to the IL-6 promoter is dramatically reduced in the RelAS311A cells compared with the controls. Consistent with the role of ζPKC in this mechanism, binding of Pol II to the IL-6 promoter is severely reduced in ζPKC–/– EFs (Figure 9C). Collectively, these findings support the notion that by directly phosphorylating RelA Ser311, ζPKC controls the recruitment of essential components of the transcriptional machinery and consequently the efficiency of κB-transcriptional activation.
Fig. 8. Role of Ser311 in CBP-enhanced NF-κB transcriptional activity. EFs stably expressing wild-type or the S311A RelA mutant were transfected with the κB-dependent reporter and the Renilla control plasmid, along with increasing amounts of a CBP expression plasmid, after which they were either untreated or stimulated with TNF-α for 6 h and the luciferase activity was determined as above (A). Immublot analysis with an anti-CBP antibody shows the levels of expression of the transfected CBP (B).
Fig. 9. RelA–CBP interaction and promoter recruitment. (A) EFs stably expressing wild-type or the S311A RelA mutant were stimulated with TNF-α for 1 h, after which cell extracts were immunoprecipitated with an anti-HA antibody and the immunoprecipitates were analyzed by immunoblotting with an anti-CBP or anti-HA antibody (left panel). Wild-type or ζPKC-deficient EFs, or ζPKC-deficient EFs in which HA-ζPKC was ectopically expressed were stimulated with TNF-α for 1 h, after which endogenous RelA was immunoprecipitated and the associated endogenous CBP was determined by immunoblotting (right panel). (B) Cells as above were stimulated or not with TNF-α for 90 min, after which cross-linked chromatin was immunoprecipitated with anti-CBP or anti-RNA Pol II antibodies, and the IL-6 promoter was detected by PCR in these samples using promoter-specific primers. (C) Either wild-type or ζPKC–/– EFs were stimulated with TNF-α for 90 min and the ChIP analysis was performed to determine Pol II recruitment as above. Input shows the starting chromatin extracts.
Discussion
The results presented here strongly suggest that Ser311 is a critical ζPKC-dependent RelA phosphorylation site, which is essential for NF-κB transcriptional activity and function, through the regulation of the interaction of RelA with the transcriptional co-activator CBP, and its recruitment to κB-dependent promoters. Other phosphorylation sites in RelA have been implicated in the regulation of NF-κB function. Not only has the transactivation domain of RelA been reported to be targeted by CKII (Ser 529) and IKKβ (Ser 536), but also compelling evidence indicates that the RHD must be phosphorylated for RelA to be transcriptionally competent. Thus, previous studies identified phosphorylation of Ser276 as a critical event for RelA transcriptional activity (Zhong et al., 1997). The kinase responsible for this phosphorylation may be PKA in lipopolysaccharide-activated 70Z/3 cells (Zhong et al., 1997, 1998) and MSK in TNF-α-activated cells (Vermeulen et al., 2003). Also, S276 has been reported to be constitutively phosphorylated in vivo (Anrather et al., 1999). What we show here is that ζPKC, which we previously had described to phosphorylate the RHD of RelA directly in vitro (Leitges et al., 2001), targets Ser311. These observations are particularly relevant since RelA phosphorylation is severely impaired in vivo in EFs from ζPKC knockout mice (Leitges et al., 2001). Here, using a specific anti-phosho(Ser311) antibody, we demonstrate that this site is phosphorylated in endogenous RelA in activated mouse fibroblasts. Furthermore, we show here that endogenous phosphorylation of this site is largely inhibited in ζPKC knockout EFs, suggesting that the role of ζPKC cannot be compensated by the presence in EFs of the highly related, and abundantly expressed, λ/ιPKC isoform. Interestingly, we also show that Ser311 phosphorylation is essential for the interaction of CBP with RelA in vivo, and for the ability of CBP to synergize with RelA for the activation of κB-dependent transcription. In addition, the loss of ζPKC severely impairs the CBP–RelA endogenous interaction. Consistent with this, we show that the recruitment of CBP to the IL-6 promoter in vivo is impaired by the S311A mutation. Interestingly, this mutation also abrogates the binding of Pol II to the IL-6 promoter, which is also inhibited in the ζPKC–/– EFs. Therefore, it seems that phosphorylation of at least two sites (Ser276 and Ser311) in the RelA RHD may drive CBP interaction and an optimal NF-κB transcriptional activity. In this regard, the constitutive phosphorylation of Ser276 (Anrather et al., 1999) or its potential induced phosphorylation by TNF-α in fibroblasts (Vermeulen et al., 2003) may help the interaction of CBP driven by S311 phosphorylation. However, our data demonstrating that both sites are independently targeted by PKA and ζPKC suggest that under some conditions, both may control NF-κB activity in response to separate pathways. Interestingly, Zhong et al. (1998) noted that although RelA and CREB belong to distinct transcription factor families, the sequence targeted by PKA (Ser133 in CREB) responsible for the recruitment of CBP shows a high degree of homology with that of RelA. Interestingly, whereas Ser276 (in RelA) and Ser133 (in CREB) lie at different ends of the homologous region, Ser311 in RelA is located in the proximity of the CREB Ser133. The implications of this observation for the detailed structural relationship controlling the formation of the RelA–CBP complex during NF-κB activation warrants further investigation. In any event, these observations are of great functional importance because they constitute a significant advance in the understanding of the signaling elements that control NF-κB transcriptional activation downstream of IKK, and establish the mechanism and importance of ζPKC in the control of this critical signaling pathway.
Materials and methods
Reagents and antibodies
TNF-α (Promega), cycloheximide (Sigma) and etoposide (Calbiochem) were purchased. Polyclonal anti-RelA (C-20), anti-CBP (A-22), anti-Pol II (N-20) and anti-HA (Y-11) were from Santa Cruz. Monoclonal anti-HA (12CA5) was from Roche. Affinity-purified phospho-Ser311-RelA polyclonal antibody was from Alpha Diagnostic International. All antibodies were used according to the manufacturers’ instructions. IL-6 levels were determined using the OptEIA ELISA kit from Becton Dickinson, following the manufacturer’s procedures.
Cell lines
293 cells and mouse EFs were grown in Dulbecco’s modified Eagle’s medium (DMEM; Gibco-BRL) supplemented with 10% (v/v) fetal calf serum (FCS; Gibco-BRL) in an atmosphere of 95% air and 5% CO2. Mouse EFs either wild-type or ζPKC–/– have been described previously (Leitges et al., 2001). RelA knockouts were a gift from Dr Baltimore (Caltec). Some cell cultures were transduced with pBabe, pBabe-RelA, pBabe-RelAS311A or pBabe-ζPKC retroviral constructs as described (Barradas et al., 1999). All mutants of RelA (obtained in GST, pBabe or pcDNA3-HA vectors) were generated by site-directed mutagenesis (Stratagene) and the sequence was fully confirmed.
In vitro phosphorylation and tryptic mapping
GST fusion proteins for RelA-RHD (amino acids 12–317), RelAT308A, RelAS311A and RelAS276A mutants were prepared following the manufacturer’s instructions (Amersham Biosciences). The RelA mutants were generated by site-directed mutagenesis (Stratagene). For in vitro phosphorylation assays, 5 µg of recombinant RelA or mutants were incubated at 30°C for 60 min in kinase assay buffer containing 25 mM Tris–HCl pH 7.5, 5 mM MgCl2, 0.5 mM EGTA, 1 mM dithiothreitol (DTT), 100 µM ATP and 50 µCi of [γ-32P]ATP in the presence of baculovirus-expressed ζPKC (Panvera). In vitro phosphorylation with PKA was performed as previously described (Zhong et al., 1998). For peptide mapping, radiolabeled proteins were resolved by SDS–PAGE and transferred to a nitrocellulose membrane. After transfer, the moistened membrane was covered with Saran Wrap and exposed to autoradiography. The protein band of interest was identified, cut out, and soaked in 0.5% polyvinylpyrrolidone in 100 mM acetic acid for 30 min at 37°C in a microcentrifuge tube. Afterwards, the piece of membrane was washed extensively with water (5 × 1 ml) and twice with freshly made 50 mM ammonium bicarbonate. Then, the membrane was incubated with 10 µg of TPCK-treated trypsin (Worthington Biochemical Corporation) in 200 µl of 50 mM ammonium bicarbonate pH 8.0 at 37°C. After 2 h, another 10 µg of TPCK-trypsin were added to the sample and incubated for another 2 h at 37°C. At the end of the reaction, 300 µl of water were added to the sample and centrifuged at 10 000 r.p.m. in a microcentrifuge for 5 min at room temperature. The supernatant (containing ∼90% of the radioactivity) was transferred to a new tube and lyophilized in a centrifugal vacuum concentrator. Once dried, peptides were oxidized in 50 µl of ice-cold performic acid for 60 min on ice. Then, 400 µl of water were added, and the sample was frozen on dry ice and lyophilized several times. Finally, the sample was dissolved in 5 µl of electrophoresis buffer (1% ammonium carbonate, pH 8.9), spotted on a TLC plate and electrophoresed on the first dimension for 30 min at 1.0 kV in pH 8.9 buffer. Separation in the second dimension was achieved by ascending chromatography in the following phospho-chromatography buffer: 37.5% n-butanol, 25% pyridine, 7.5% glacial acetic acid. After chromatography (12–14 h), the plates were dried in a fume hood, covered with Saran Wrap and exposed to X-ray film using an intensifier screen.
Immunoprecipitation and western blot assays
Cells plated on 10 cm diameter dishes were stimulated or not with 10 ng/ml of TNF-α for different times, after which cells were lysed with cell extraction buffer (Cell Signaling) for detection of RelA phosphorylation or with the following buffer for the CBP interaction assays: 25 mM HEPES–KOH pH 7.2, 150 mM potassium acetate, 2 mM EDTA, 0.1% NP-40, 10 mM sodium fluoride, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride (PMSF) and protease inhibitors. Soluble fractions were immunoprecipitated for 16 h at 4°C with the indicated antibodies. The immunoprecipitates were washed extensively, separated by SDS–PAGE and transferred to nitrocellulose-ECL membranes (Amersham Biosciences). The membranes were incubated with the appropriate antibodies, washed, and incubated with horseradish peroxidase-coupled secondary antibodies. After washing, the blots were visualized by using ECL reagent (Amersham Biosciences).
Chromatin immunoprecipitation analysis
Following stimulation, EFs were fixed by adding directly to the culture medium formaldehyde (HCHO; from a 37% HCHO–10% methanol stock, Calbiochem) to a final concentration of 1%. After 5 min, ice-cold phosphate-buffered saline (PBS) was added, and cells were rapidly collected, centrifuged and washed three times (10 min each) with PBS. Cells were lysed for 5 min in L1 buffer (50 mM Tris pH 8.0, 2 mM EDTA, 0.1% NP-40 and 10% glycerol) supplemented with protease inhibitors. Nuclei were pelleted at 3000 r.p.m. and resuspended in L2 buffer (50 mM Tris pH 8.0, 0.1% SDS and 5 mM EDTA). Chromatin was sheared by sonication, centrifuged and diluted 10 times in dilution buffer (50 mM Tris pH 8.0, 0.5% NP-40, 0.2 M NaCl and 0.5 mM EDTA). Extracts were pre-cleared for 3 h with 60 µl of a 50% suspension of salmon sperm-saturated protein A–agarose. Immunoprecipitations were carried out overnight at 4°C. Immunocomplexes were collected with salmon sperm-saturated protein A for 30 min and washed three times (5 min each) with high-salt buffer (20 mM Tris pH 8.0, 0.1% SDS, 1% NP-40, 2 mM EDTA and 0.5 M NaCl) followed by three washes in no salt buffer (1× TE). Immunocomplexes were extracted in 1× TE containing 2% SDS, and protein–DNA cross-links were reverted by heating at 65°C overnight. After proteinase K digestion, DNA was extracted with phenol–chloroform and precipitated in ethanol. About one-twentieth of the immunoprecipitated DNA was used in each PCR. The following promoter-specific primers were used: IL-6, sense 5′-GACATG CTCAAGTGCTGAGTCAC-3′; antisense 5′-AGATTGCACAATGTG ACGTCG-3′. The PCR conditions were as follows: 1.25 U of Taq DNA polymerase (Amersham Biosciences), 100 ng of each primer, 200 µM dNTP, 2.5 µl of 10× Taq buffer and double-distilled water to a final volume of 25 µl: 94°C for 180 s; 34 cycles at 94°C for 45 s, 60°C for 60 s and 72°C for 60 s; final elongation at 72°C for 10 min.
Apoptosis assay
Subconfluent mouse EFs transduced with the different retroviral constructs were incubated with TNF-α (10 ng/ml) and cycloheximide (2 µg/ml) or etoposide (100 µM) for 24 or 72 h, after which the percentage of apoptotic cells was determined by TUNEL using the Mebstain Apoptosis Kit II (MBL International).
Luciferase assays
Subconfluent cultures were transfected with Lipofectamine Plus (Invitrogen) with 100 ng of a multimerized κB-luciferase reporter gene plasmid (described previously in Leitges et al., 2001) and 2 ng of the Renilla control reporter pRL-CMV (Promega). In some experiments, cells were co-transfected with different amounts of pCDNA3-HA-RelA, pCDNA3-HA-RelAS311A or pCDNA3-HA-CBP. After 24 h, cells were stimulated or not for 6 h with different concentrations of TNF-α, IL-1β or ACH6, extracts were prepared and the levels of luciferase activity were determined using the Dual-Luciferase Reporter Assay System according to the manufacturer’s instructions (Promega).
Electrophoretic mobility shift assay
EMSA experiments for NF-κB were performed as previously described (Leitges et al., 2001).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Dr G.Natoli for help and advice in the ChIP assays. J.M. is a recipient of the Ayuda Investigación Juan March 2001. This work was supported by grants SAF1999-0053 (to M.T.D.-M.) and SAF2002-0187 (to J.M.) from MCYT, and by an institutional grant from Fundación Ramón Areces to the CBM. A.D. is an MCYT fellow.
References
- Anrather J., Csizmadia,V., Soares,M.P. and Winkler,H. (1999) Regulation of NF-κB RelA phosphorylation and transcriptional activity by p21(ras) and protein kinase Cζ in primary endothelial cells. J. Biol. Chem., 274, 13594–13603. [DOI] [PubMed] [Google Scholar]
- Barradas M., Monjas,A., Diaz-Meco,M.T., Serrano,M. and Moscat,J. (1999) The downregulation of the pro-apoptotic protein Par-4 is critical for Ras-induced survival and tumor progression. EMBO J., 18, 6362–6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnard M. et al. (2000) Deficiency of T2K leads to apoptotic liver degeneration and impaired NF-κB-dependent gene transcription. EMBO J., 19, 4976–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudio E., Brown,K., Park,S., Wang,H. and Siebenlist,U. (2002) BAFF-induced NEMO-independent processing of NF-κB2 in maturing B cells. Nat. Immunol., 3, 958–965. [DOI] [PubMed] [Google Scholar]
- Cowley S., Paterson,H., Kemp,P. and Marshall,C.J. (1994) Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell, 77, 841–852. [DOI] [PubMed] [Google Scholar]
- Dejardin E. et al. (2002) The lymphotoxin-β receptor induces different patterns of gene expression via two NF-κB pathways. Immunity, 17, 525–535. [DOI] [PubMed] [Google Scholar]
- Ghosh S. and Karin,M. (2002) Missing pieces in the NF-κB puzzle. Cell, 109, S81–S96. [DOI] [PubMed] [Google Scholar]
- Hoeflich K.P., Luo,J., Rubie,E.A., Tsao,M.S., Jin,O. and Woodgett,J.R. (2000) Requirement for glycogen synthase kinase-3β in cell survival and NF-κB activation. Nature, 406, 86–90. [DOI] [PubMed] [Google Scholar]
- Karin M. and Lin,A. (2002) NF-κB at the crossroads of life and death. Nat. Immunol., 3, 221–227. [DOI] [PubMed] [Google Scholar]
- Karin M., Cao,Y., Greten,F.R. and Li,Z.W. (2002) NFκB in cancer: from innocent bystander to major culprit. Nat. Rev. Cancer, 2, 301–310. [DOI] [PubMed] [Google Scholar]
- Kayagaki N. et al. (2002) BAFF/BLyS receptor 3 binds the B cell survival factor BAFF ligand through a discrete surface loop and promotes processing of NF-κB2. Immunity, 17, 515–524. [DOI] [PubMed] [Google Scholar]
- Lallena M.J., Diaz-Meco,M.T., Bren,G., Pay,C.V. and Moscat,J. (1999) Activation of IκB kinase β by protein kinase C isoforms. Mol. Cell. Biol., 19, 2180–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitges M. et al. (2001) Targeted disruption of the ζPKC gene results in the impairment of the NF-κB pathway. Mol. Cell, 8, 771–780. [DOI] [PubMed] [Google Scholar]
- Li Q. and Verma,I.M. (2002) NF-κB regulation in the immune system. Nat. Rev. Immunol., 2, 725–734. [DOI] [PubMed] [Google Scholar]
- Madrid L.V., Wang,C.-Y., Guttridge,D.C., Schottelius,A.J.G., Baldwin,A.S.,Jr and Mayo,M.W. (2000) Akt suppresses apoptosis by stimulating the transactivation potential of the RelA/p65 subunit of NF-κB. Mol. Cell. Biol., 20, 1626–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P., Duran,A., Minguet,S., Gaspar,M.L., Diaz-Meco,M.T., Rennert,P., Leitges,M. and Moscat,J. (2002) Role of ζPKC in B-cell signaling and function. EMBO J., 21, 4049–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J. and Diaz-Meco,M.T. (2000) The atypical protein kinase Cs. Functional specificity mediated by specific protein adapters. EMBO Rep., 1, 399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J., Diaz-Meco,M.T. and Rennert,P. (2003) NF-κB activation by protein kinase C isoforms and B-cell function. EMBO Rep., 4, 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai H., Chiba,H., Miyoshi,H., Sugita,T. and Toriumi,W. (1999) IκB kinases phosphorylate NF-κB p65 subunit on serine 536 in the transactivation domain. J. Biol. Chem., 274, 30353–30356. [DOI] [PubMed] [Google Scholar]
- Senftleben U. et al. (2001) Activation by IKKα of a second, evolutionarily conserved, NF-κB signaling pathway. Science, 293, 1495–1499. [DOI] [PubMed] [Google Scholar]
- Sizemore N., Leung,S. and Stark,G.R. (1999) Activation of phosphatidylinositol 3-kinase in response to interleukin-1 leads to phosphorylation and activation of the NF-κB p65/RelA subunit. Mol. Cell. Biol., 19, 4798–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G., Minemoto,Y., Dibling,B., Purcell,N.H., Li,Z., Karin,M. and Lin,A. (2001) Inhibition of JNK activation through NF-κB target genes. Nature, 414, 313–317. [DOI] [PubMed] [Google Scholar]
- Vermeulen L., De Wilde,G., Damme,P.V., Vanden Berghe,W. and Haegeman,G. (2003) Transcriptional activation of the NF-κB p65 subunit by mitogen- and stress-activated protein kinase-1 (MSK1). EMBO J., 22, 1313–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. and Baldwin,A.S.,Jr (1998) Activation of nuclear factor-κB-dependent transcription by tumor necrosis factor- α is mediated through phosphorylation of RelA/p65 on serine 529. J. Biol. Chem., 273, 29411–29416. [DOI] [PubMed] [Google Scholar]
- Wang D., Westerheide,S.D., Hanson,J.L. and Baldwin,A.S.,Jr (2000) Tumor necrosis factor α-induced phosphorylation of RelA/p65 on Ser529 is controlled by casein kinase II. J. Biol. Chem., 275, 32592–32597. [DOI] [PubMed] [Google Scholar]
- Xiao G., Harhaj,E.W. and Sun,S.C. (2001) NF-κB-inducing kinase regulates the processing of NF-κB2 p100. Mol. Cell, 7, 401–409. [DOI] [PubMed] [Google Scholar]
- Zhong H., SuYang,H., Erdjument-Bromage,H., Tempst,P. and Ghosh,S. (1997) The transcriptional activity of NF-κB is regulated by the IκB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell, 89, 413–424. [DOI] [PubMed] [Google Scholar]
- Zhong H., Voll,R.E. and Ghosh,S. (1998) Phosphorylation of NF-κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell, 1, 661–671. [DOI] [PubMed] [Google Scholar]