Abstract
The nonsense-mediated mRNA decay (NMD) pathway promotes the rapid degradation of mRNAs containing premature stop codons (PTCs). In Caenorhabditis elegans, seven genes (smg1–7) playing an essential role in NMD have been identified. Only SMG2–4 (known as UPF1–3) have orthologs in Saccharomyces cerevisiae. Here we show that the Drosophila orthologs of UPF1–3, SMG1, SMG5 and SMG6 are required for the degradation of PTC-containing mRNAs, but that there is no SMG7 ortholog in this organism. In contrast, orthologs of SMG5–7 are encoded by the human genome and all three are required for NMD. In human cells, exon boundaries have been shown to play a critical role in defining PTCs. This role is mediated by components of the exon junction complex (EJC). Contrary to expectation, however, we show that the components of the EJC are dispensable for NMD in Drosophila cells. Consistently, PTC definition occurs independently of exon boundaries in Drosophila. Our findings reveal that despite conservation of the NMD machinery, different mechanisms have evolved to discriminate premature from natural stop codons in metazoa.
Keywords: EJC/mRNA decay/NMD/RNPS1/Y14
Introduction
Nonsense (codon)-mediated mRNA decay (NMD) is an RNA surveillance pathway that degrades mRNAs possessing premature translation termination codons (PTCs) (reviewed in Wagner and Lykke-Andersen, 2002). In mammalian cells, NMD is linked to splicing. Indeed, the position of a stop codon relative to the 3′-most intron has emerged as a critical determinant of whether it will be interpreted as being premature or natural (reviewed in Nagy and Maquat, 1998). Natural stop codons are typically located within the terminal exon, while destabilizing nonsense codons lie >50 nucleotides upstream of the last exon–exon boundary. This observation gave rise to the hypothesis that the process of splicing deposits a mark at exon–exon junctions. These marks are used to discriminate premature stops (upstream of the last mark) from natural stops (downstream of the last mark) (reviewed in Nagy and Maquat, 1998; Wagner and Lykke-Andersen, 2002).
The exon junction complex (EJC) is thought to provide the splicing marks for NMD. This multiprotein complex binds ∼20–24 nucleotides upstream of splice junctions in a splicing-dependent manner (Le Hir et al., 2001). Although the precise biochemical composition of the EJC remains to be established, at least five proteins have been identified as components of the EJC assembled in vitro. These include REF1 (also known as Aly), SRm160, RNPS1 and Y14:MAGOH heterodimers (Le Hir et al., 2001). Y14:MAGOH dimers, and possibly other EJC proteins, remain bound to the newly exported mRNAs and could be used in the cytoplasm to mark the location of exon–exon boundaries (Kim et al., 2001a,b; Le Hir et al., 2001; Lykke-Andersen et al., 2001). Consistently, both RNPS1 and Y14:MAGOH have been shown to elicit NMD when tethered downstream of a stop codon (Lykke-Andersen et al., 2001; Fribourg et al., 2003; Gehring et al., 2003).
In Saccharomyces cerevisiae, most genes lack introns and, apart from REF, none of the known components of the EJC is conserved. Consistently, exon boundaries are not used to distinguish natural from premature stop codons in budding yeast. Instead, yeast mRNAs are marked by loosely defined downstream sequence elements (DSEs) that might have a function analogous to that of mammalian exon junctions (reviewed in Wagner and Lykke-Andersen, 2002).
The genetics of NMD have been studied in S.cerevisiae and Caenorhabditis elegans. Three yeast (UPF1–3) and seven C.elegans genes (smg1–7, suppressor with morphogenetic effects on genitalia) that play an essential role in NMD have been identified. The UPF1–3 proteins (also known as SMG2–4 in C.elegans) are conserved from yeast to humans, while SMG1 has orthologs in higher eukaryotes but not in S.cerevisiae (reviewed in Wagner and Lykke-Andersen, 2002).
In mammalian cells, UPF1 is a nucleocytoplasmic shuttling protein with a predominantly cytoplasmic localization at equilibrium (Lykke-Andersen et al., 2000; Mendell et al., 2002). UPF2 localizes around the nucleus and is known to bind UPF3 and UPF1 (He et al., 1997; Lykke-Andersen et al., 2000). The two human UPF3 proteins (UPF3a and UPF3b) are found predominantly within the nucleus and associate preferentially with spliced RNAs, possibly through interactions with RNPS1 and/or Y14:MAGOH (Lykke-Andersen et al., 2000, 2001; Kim et al., 2001b; Le Hir et al., 2001; Serin et al., 2001; Gehring et al., 2003). The interactions of the translation termination factors eRF1 and eRF3 with UPF1 and of eRF3 with UPF2 and UPF3 provide the link between NMD and translation termination (reviewed in Wagner and Lykke-Andersen, 2002).
The smg-1 gene encodes a phosphoinositide-3-kinase-related protein kinase required for the phosphorylation of UPF1 (Page et al., 1999; Denning et al., 2001; Pal et al., 2001; Yamashita et al., 2001), while the SMG5 and SMG7 proteins function in the dephosphorylation of UPF1 (Page et al., 1999; Anders et al., 2003; Chiu et al., 2003). Both SMG5 and SMG7 are characterized by the presence of one and a half N-terminal tetratricopeptide repeats (TPRs) (Cali et al., 1999; Clissold and Ponting, 2000). In addition, SMG5 has a C-terminal PIN (PilT-N-terminal) domain which is also present in several proteins possessing exonuclease activity (Clissold and Ponting, 2000).
Current data support a model in which mammalian UPF3 is loaded onto mRNAs during splicing via interactions with components of the EJC, while UPF2 joins the complex soon after export. UPF3 and UPF2 are displaced by the ribosomes as they traverse the mRNA during the first round of translation. Pausing of translating ribosomes at a premature stop codon would lead to the recruitment of UPF1, most probably via interactions with the eRF1–eRF3 complex, and to the incomplete removal of UPF3 and/or UPF2 proteins from downstream mRNA sequences. This event creates an opportunity for the assembly of the NMD complex, consisting of at least UPF1, UPF2 and UPF3. The mRNA serving as a platform for the assembly of this complex is then targeted for rapid degradation (reviewed in Wagner and Lykke-Andersen, 2002).
Although the NMD pathway is becoming increasingly well understood in yeast, the molecular mechanism of NMD in higher eukaryotes remains unclear. In particular, the degradation route followed by aberrant mRNAs in metazoa is unknown. Moreover, the yeast pathway differs from higher eukaryotic NMD in several aspects. First, phosphorylation of Upf1p does not appear to occur in yeast. Secondly, there are no obvious SMG5 or SMG7 orthologs encoded by the yeast genome. Thirdly, in yeast, premature stop codons are defined independently of exon boundaries. Since most genes in higher eukaryotes have introns, and components of the EJC are conserved, it is assumed that exon boundaries would play a critical role in defining premature stop codons in metazoa.
In order to shed light on the NMD pathway in higher eukaryotes, we have generated Drosophila cell lines constitutively expressing PTC-containing mRNAs. Using double-stranded RNA interference (RNAi), we show that the Drosophila orthologs of UPF1, UPF2, UPF3 and SMG1 are required for degradation of the NMD reporters. Database searches reveal that orthologs of the C.elegans proteins SMG5–7 are encoded by all animal genomes sequenced so far, but not by fungi, indicating that SMG5–7 are a specific feature of the NMD pathway in higher eukaryotes. Interestingly, the Drosophila genome encodes orthologs of SMG5 and SMG6, but not of SMG7. We show that SMG5 and SMG6 are both required for NMD in Drosophila cells. Unexpectedly, the definition of PTCs in Drosophila occurs by a novel mechanism that is independent of exon boundaries and may not rely on specific DSEs. Our findings reveal that despite conservation of the NMD machinery, different mechanisms have evolved to discriminate premature from natural stop codons in metazoa.
Results
Generation of an NMD reporter for Drosophila cells
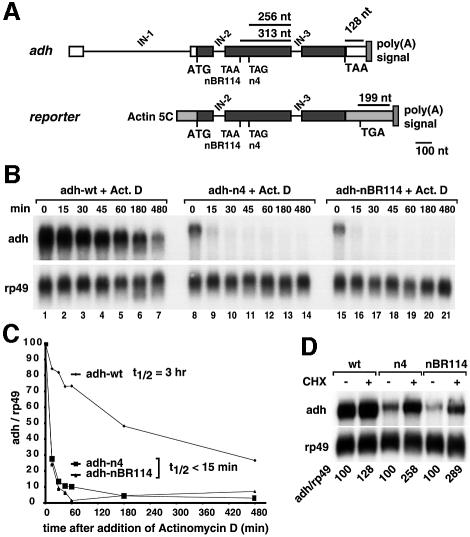
Mutations in the alcohol dehydrogenase gene (adh) of Drosophila that generate PTCs lead to reduced levels of cytoplasmic and nuclear mRNA, and to mRNAs with poly(A) tails longer than the wild-type transcript (Brogna, 1999). To investigate whether adh alleles carrying PTCs are degraded by the NMD pathway and to gain further insight into the mechanism of this pathway in higher eukaryotic cells, we designed a reporter gene in which the genomic coding region of Drosophila adh is placed downstream of the constitutive actin 5C promoter and upstream of a polyadenylation site derived from SV40 (Figure 1A). In adult flies, transcription of the adh gene produces a pre-mRNA with three introns (Figure 1A). The first intron (IN-1) is located in the 5′-untranslated region (5′-UTR), while introns 2 and 3 (IN-2 and IN-3) lie between codons 33 and 34, and 168 and 169, respectively. The complete coding sequence has 257 codons (Brogna, 1999). In the reporter constructs used in this study, the adh 5′- and 3′-UTRs were deleted (Figure 1A).
Fig. 1. Generation of an NMD reporter for Drosophila cells. (A) Schematic representation of the adh gene and the reporter used in this study. Black boxes, exons; white boxes, adh 5′- or 3′-UTRs; gray boxes, sequences derived from vector pAc5.1b; IN, introns. The PTC-containing constructs correspond to adh-nBR114 and adh-n4 (Brogna, 1999). (B) S2 cells constitutively expressing adh-wt or the PTC reporters were incubated with actinomycin D (5 µg/ml) for the times indicated above the lanes. Total RNA samples were isolated and analyzed by northern blot using probes specific for adh and rp49 mRNAs. (C) The levels of adh-wt, adh-n4 and adh-nBR114 were quantitated, normalized to the levels of rp49 mRNA (whose levels do not change relative to 18S rRNA within the time frame of the experiment) and plotted as a function of time. The half-lives of the mRNAs are indicated. (D) S2 cells constitutively expressing adh-wt or the PTC reporters were incubated with cycloheximide (CHX, 100 µg/ml) for 45 min. Total RNA samples were analyzed by northern blot. The numbers below the lanes indicate the levels of adh transcripts normalized to those of rp49 mRNA. These values were set to 100% in untreated cells.
Two previously characterized PTCs were introduced in the adh reporter by site-directed mutagenesis. These correspond to adh-nBR114 and adh-n4, which carry PTCs at codons 64 and 83, respectively (Brogna, 1999). The reporter constructs were co-transfected with a plasmid encoding puromycin acetyl transferase into Drosophila Schneider cells (S2 cells) to generate polyclonal cell lines constitutively expressing wild-type adh (adh-wt) or one of the two PTC mutants. The steady-state levels of the corresponding transcripts were analyzed by northern blot. In agreement with previous observations (Brogna, 1999), adh-wt mRNA had a higher electrophoretic mobility and was expressed at higher levels than the adh-n4 and adh-nBR114 mRNAs (Figure 1B, lanes 1, 8 and 15). In all experiments performed in Drosophila cells, the levels of the NMD reporters were normalized to those of the endogenous rp49 mRNA (encoding ribosomal protein L32), which is a long-lived mRNA (half-life >8 h). Following this normalization, the levels of PTC-containing transcripts were reduced to 25–30% of wild-type adh levels.
We next determined the stabilities of the adh transcripts. In the experiments shown in Figure 1B, transcription was inhibited by actinomycin D, cells were harvested at the indicated time points and the adh mRNA levels were determined. The half-life of adh-wt was ∼3 h, while the half-lives of nBR114 or n4 mRNAs were <15 min (Figure 1B and C). Thus, the overall reduction of the steady-state levels of the PTC-containing mRNAs is a direct consequence of their higher turnover rate.
mRNAs degraded through the NMD pathway are stabilized following inhibition of translation (see Zhang et al., 1998b). When cells constitutively expressing the adh reporters were treated with cycloheximide for 45 min, the steady-state levels of n4 and nBR114 mRNAs were increased 2.5- and 2.8-fold, respectively (Figure 1D).
To assess definitively whether adh-nBR114 and adh-n4 were subjected to NMD, we investigated the effect of depleting endogenous UPF1 or UPF2 on the expression levels of these mRNA species. S2 cells expressing adh-wt or the PTC mutants were transfected with dsRNAs specific for Dm UPF1 or Dm UPF2. To ensure that the proteins were depleted efficiently, all RNAi experiments described in this study were carried out by transfecting cells twice with the corresponding dsRNAs, with the second transfection 4 days after the first on day 0. The expression levels of the adh mRNAs were analyzed on day 7. Depletion of either UPF1 or UPF2 resulted in a 4- to 5-fold increase of adh-n4 or adh-nBR114 mRNA levels (Figure 2B and C). The expression levels of the wild-type mRNA were not significantly changed (Figure 2A). Together, these results indicate that adh-n4 and nBR114 mRNAs are degraded through the NMD pathway.
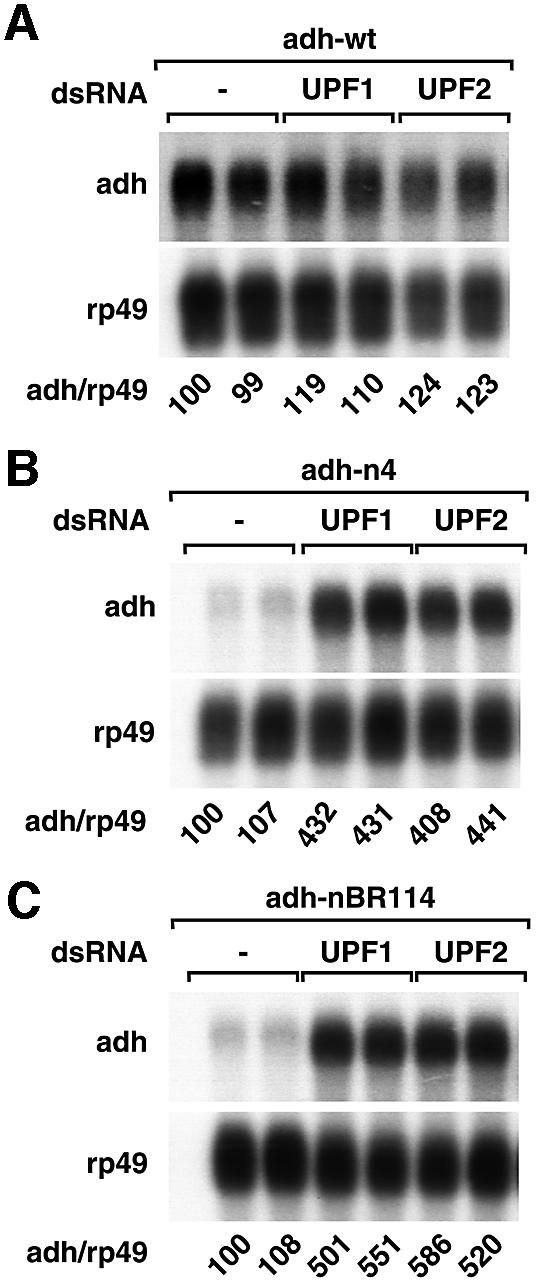
Fig. 2. Adh reporters are subjected to NMD in Drosophila cells. (A–C) S2 cell lines expressing adh-wt, adh-n4 or adh-nBR114 were transfected with dsRNAs corresponding to UPF1 or UPF2 as indicated. Transfections were carried out in duplicate. Total RNA samples were analyzed by northern blot as described in Figure 1.
Orthologs of SMG5–7 are encoded by all sequenced animal genomes
Having established a reporter assay that reproduces bona fide NMD, we next investigated whether the Drosophila orthologs of UPF3, SMG1 and SMG5–7 are involved in this process. Using BLAST searches (Altschul et al., 1997), we identified single genes encoding SMG1 and UPF3 in Drosophila genomic sequences (Adams et al., 2000). Furthermore, the products of the Drosophila (Dm) genes CG8954 (anon-34Ea) and CG6369 were reported to be related to C.elegans (Ce) SMG7 and SMG5 (Clissold and Ponting, 2000; Chiu et al., 2003). Dm CG8954, Dm CG6369, Ce SMG5 and Ce SMG7 are characterized by the presence of one and a half N-terminal TPRs. With the exception of Ce SMG7, these proteins have, in addition, a C-terminal PIN domain (Clissold and Ponting, 2000).
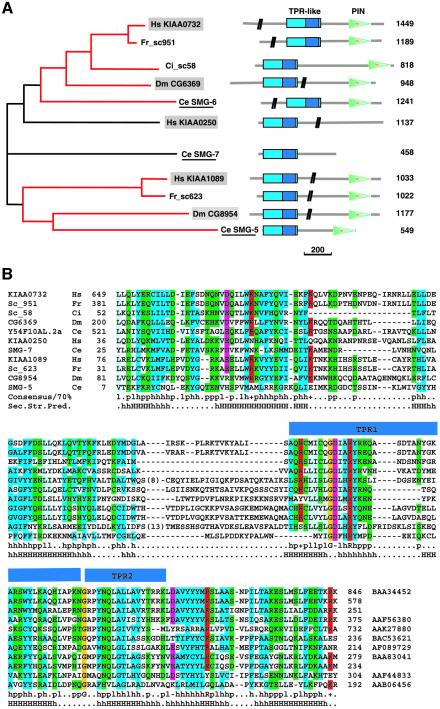
The orthologous relationships between these proteins had not been assigned in previous studies (Clissold and Ponting, 2000; Chiu et al., 2003); we therefore carried out database searches in order to retrieve from the completely sequenced genomes all proteins with a TPR-like repeat at the N-terminus and a PIN domain at the C-terminus. A multialignment of the retrieved sequences revealed that the conserved region of ∼50 residues encompassing the identified TPR repeats (Clissold and Ponting, 2000) could be expanded considerably (Figure 3A and B) and was predicted to be entirely helical over a length of ∼150 residues (Figure 3B).
Fig. 3. Phylogenetic tree and domain organization of the SMG5–7 proteins. (A) The two identified orthologous groups are shown in red. Only one and a half TPR repeats can be identified automatically and are indicated in dark blue within the conserved helical N-terminal domain (cyan). The C.elegans sequences are underlined and are known to be involved in NMD (Cali et al., 1999; Page et al., 1999; Anders et al., 2003). Human and Drosophila sequences characterized in this study are shown in gray boxes. The vertical black bars on the schematic of the protein domains indicate that part of the protein sequence is not drawn to scale. The number of amino acid residues is shown on the right. Scale bar: 200 amino acids. TPR, tetratricopeptide repeats; PIN, PilT-N-terminus; Ce, Caenorhabditis elegans; Ci, Ciona intestinalis; Dm, Drosophila melanogaster; Fr, Fugu rubripes; Hs, Homo sapiens. (B) Multiple sequence alignment of the TPR-like region. The accession number of each sequence is indicated, as well as the species and the start and end points of the sequences. For the sequences of C.intestinalis and F.rubripes, no accession numbers are shown, as the predictions were performed directly on the genomic sequences using GeneWise (Birney et al., 1996). The consensus in 70% of the sequences is shown below the multialignment; h, l, p, +, – represent hydrophobic, aliphatic, polar, positive and negative residues, respectively. Aliphatic residues are highlighted in cyan, hydrophobic in blue, polar in green, negative in pink, and positive in red. The conserved glycines are highlighted in orange. The secondary structure prediction (Sec.Str.Pred.) is taken from the consensus of the multiple alignment (H, helices predicted with an accuracy >82%; h, helices predicted with an accuracy ≤82%]. The blue bars above the sequences show the position of the one and a half TPR repeats identified by Clissold and Ponting (2000).
Using the expanded TPR-like helical region as a query, two more proteins were retrieved from the database that did not contain a PIN domain: Ce SMG7 and Hs KIAA0250. A multiple alignment of the family revealed two distinct orthologous groups across the species considered: one includes Ce SMG5, Hs KIAA1089 and Dm CG8954, and the other Hs KIAA0732, Dm CG6369 and an additional Ce protein encoded by the Y54F10AL-2a region. This protein corresponds to Ce SMG6 (P.Anderson, personal communication). Only for the proteins without PIN domains could the orthologous relationships not be assigned. The phylogenetic tree obtained for the N-terminal regions was identical to that obtained by using the PIN domains (data not shown).
Based on the domain organization and on the functional characterization described below, we conclude that KIAA0250 is likely to represent the human ortholog of SMG7, but that there is no SMG7 ortholog in Drosophila. As the phylogenetic tree with the PIN domain indicates that KIAA1089 and CG8954 are closely related to Ce SMG5, we propose to call these proteins Hs and Dm SMG5. KIAA0732, CG6369 and Y54F10AL-2 define another branch of the family, and we propose to call them Hs, Dm and Ce SMG6.
The Drosophila orthologs of UPF3, SMG1, SMG5 and SMG6 are essential for NMD
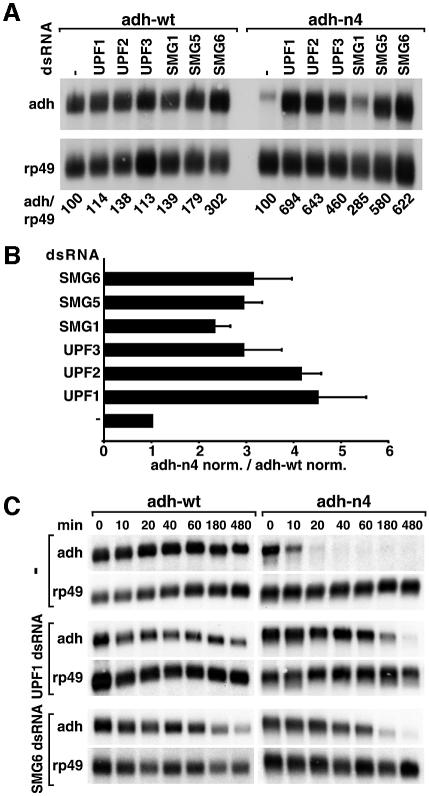
To assess whether Dm UPF3, SMG1, SMG5 and SMG6 are involved in NMD, we silenced the expression of these genes by RNAi in cells expressing adh-wt or adh-n4 transcripts. The effect of these depletions on the mRNA levels was analyzed by northern blot and compared with the effects observed when UPF1 or UPF2 were depleted. Adh-n4 mRNA levels increased 2.8- to 6-fold in cells depleted of UPF3, SMG1, SMG5 or SMG6 (Figure 4A).
Fig. 4. SMG1, SMG5 and SMG6 are essential for NMD in Drosophila cells. (A) S2 cell lines expressing adh-wt or adh-n4 were transfected with the dsRNAs indicated above the lanes. RNA samples were analyzed by northern blot, as described in Figure 2. (B) The steady-state levels of adh-wt and adh-n4 mRNAs were quantitated in at least three independent experiments and normalized to those of rp49 mRNA. For each experiment, the normalized values (norm.) obtained for n4 mRNA were divided by those obtained for adh-wt, to correct for potential non-specific effects of the depletions. This ratio was set arbitrarily to a value of 1 in control cells. The mean values ± SDs are shown. (C) S2 cell lines expressing adh-wt or adh-n4 were transfected with dsRNAs specific for UPF1 and SMG6. Seven days after addition of dsRNAs, treated and untreated cells were incubated with actinomycin D (5 µg/ml) for the times indicated above the lanes. Total RNA samples were isolated and analyzed by northern blot as described in Figure 1D.
The steady-state levels of adh-wt and adh-n4 mRNAs were quantitated in at least three independent experiments and normalized to those of rp49 mRNA. The normalized values obtained for the PTC-containing mRNA were divided by those obtained for adh-wt in order to correct for potential non-specific effects of the depletions. This ratio was set arbitrarily to a value of 1 in control cells. The up-regulation of adh-n4 mRNA was within the 2- to 4.5-fold range (Figure 4B). Similar values were reported for PTC-containing transcripts in human cells depleted of UPF1 or Y14 (Mendell et al., 2002; Gehring et al., 2003).
Because the steady-state levels of wild-type adh mRNA also increased in SMG6-depleted cells (Figure 4A), it was important to confirm that the increase in adh-n4 mRNA levels was due to a specific inhibition of the NMD pathway. We therefore tested whether the half-life of adh-n4 mRNA was specifically increased in cells depleted of SMG6. As a positive control, we measured the stability of this transcript in cells depleted of UPF1. The half-life of adh-n4 mRNA increased from <15 min in control cells to 100 min in UPF1- or SMG6-depleted cells (Figure 4C), indicating that the overall increase of the steady-state levels of this mRNA reflects a decrease of its turnover rate. The half-life of adh-wt was ∼3 h both in control cells and in cells depleted of UPF1 or SMG6 (Figure 4C). These results indicate that SMG1, SMG5 and SMG6 are essential for NMD in Drosophila.
Depletion of SMG5–7 in human cells leads to the stabilization of PTC-containing mRNAs
Although Hs SMG6 (KIAA0732) has been shown to function in the dephosphorylation of UPF1 (Chiu et al., 2003), evidence for a role for Hs SMG6 in telomere maintenance has now been reported (Reichenbach et al., 2003), raising the possibility that not all orthologs of SMG5–7 play a role in NMD. We therefore investigated whether Hs SMG5–7 are directly implicated in this process by analyzing the effect of depleting these proteins on the steady-state levels of a β-globin mRNA carrying a PTC at codon 39 (NS39; Thermann et al., 1998).
HeLa cells were transfected with short interfering RNAs (siRNAs) specific for SMG5, SMG6 or SMG7. An siRNA specific for UPF1 served as a positive control. After 48 h, cells were retransfected with the same siRNAs and plasmids expressing either wild-type β-globin or β-globin-NS39 mRNA. A plasmid expressing a green fluorescent protein (GFP) fusion (GFP–NXF1) served as a transfection control (Figure 5). The levels of the mRNAs targeted by RNAi were reduced to 30% of the levels in control cells, as estimated by RT–PCR (data not shown).
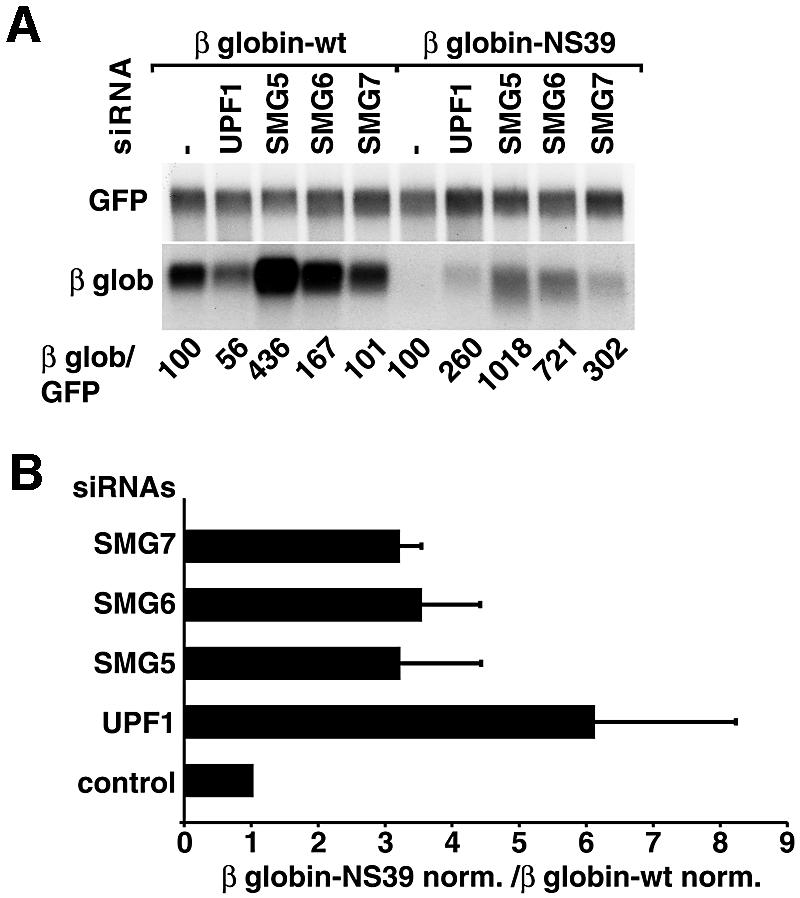
Fig. 5. Human SMG5–7 are required for NMD in HeLa cells. (A) HeLa cells were transfected with the indicated siRNAs and plasmids expressing either β-globin-wt or β-globin-NS39 mRNAs. A plasmid expressing GFP–NXF1 was co-transfected to correct for differences in transfection efficiencies. RNA samples were analyzed by northern blot using β-globin and GFP probes. The numbers below the lanes indicate the levels of β-globin-wt or NS39 transcripts normalized to those of GFP–NXF1 mRNA and set to 100% in cells treated with an unrelated siRNA (–). (B) The steady-state levels of β-globin-wt or NS39 mRNAs were quantitated in at least three independent experiments and normalized to those of the GFP–NXF1 mRNA. The normalized values (norm.) obtained for NS39 were divided by those obtained for the wild-type mRNA. The mean values ± SDs are shown.
Normalized to the levels of the transfection control, the levels of the NS39 transcript were reduced in abundance to ∼10% of the wild-type β-globin levels, as reported before (Thermann et al., 1998; Zhang et al., 1998b). Depletion of UPF1, SMG5, SMG6 or SMG7 resulted in a 2.6- to 10-fold increase of NS39 mRNA levels (Figure 5A). The expression levels of the wild-type mRNA were not significantly changed in cells depleted of SMG6 or SMG7, but were consistently decreased in UPF1- and increased in SMG5-depleted cells (Figure 5A, and data not shown). After correction for these non-specific effects, the increase in NS39 mRNA levels after UPF1, SMG5, SMG6 or SMG7 depletion was 3- to 4-fold (Figure 5B). These values are in agreement with those obtained for this and other NMD reporters in human cells depleted of UPF1 or Y14 (Mendell et al., 2002; Gehring et al., 2003). These results provide evidence for a role for human SMG5–7 in NMD.
The Drosophila orthologs of all known components of the vertebrate EJC are dispensable for NMD
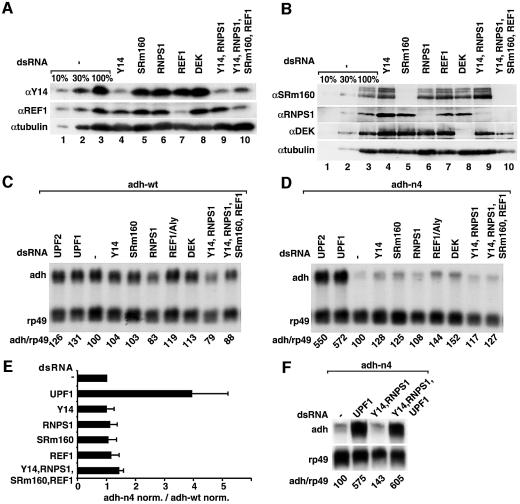
The conservation of the NMD machinery in higher eukaryotes prompted us to investigate whether the mechanism by which PTCs are defined is also conserved. To this end, we first investigated whether Drosophila EJC proteins, and in particular Y14 and RNPS1, are implicated in this process. Although it has not yet been demonstrated that the EJC is assembled in Drosophila, orthologs of all known components of the vertebrate EJC are present in this organism (Gatfield and Izaurralde, 2002). S2 cells expressing adh-wt or adh-n4 were treated with dsRNAs corresponding to the coding sequences of all known EJC proteins (i.e. REF1, RNPS1, SRm160 and Y14). The protein DEK was also depleted, although recent studies suggest that DEK may not be a bona fide component of the EJC (Gatfield and Izaurralde, 2002). As positive controls, dsRNAs corresponding to UPF1 and UPF2 were transfected. Because the EJC proteins might have partially redundant functions, we also simultaneously depleted Y14 and RNPS1 or, in addition, REF1 and SRm160. Western blot analysis revealed that 7 days after addition of dsRNAs, the expression levels of EJC proteins were ≤10% of the endogenous levels when they were depleted individually (Figure 6A and B, lanes 4–8). The depletion efficiency was reduced when more than one gene was silenced simultaneously (Figure 6A and B, lanes 9 and 10).
Fig. 6. The individual components of the EJC are dispensable for NMD in Drosophila. (A and B) S2 cells expressing adh-wt or adh-n4 were transfected with dsRNA specific for Y14, RNPS1, REF1, SRm160 or DEK, or with a mixture of dsRNAs (lanes 9 and 10) as indicated. Protein samples from total lysates of untreated or depleted cells (adh-n4 cell line) were analyzed by western blot using antibodies described in Gatfield and Izaurralde (2002). The anti-SRm160 antibody cross-reacts with three bands that are all depleted in cells treated with SRm160 dsRNA, suggesting that these bands may represent modified and/or alternatively spliced forms of the protein. Tubulin served as a loading control. In lanes 1–3, dilutions of the samples isolated on day 0 were loaded to estimate the efficiency of the depletion. (C and D) RNA samples isolated from depleted cells shown in (A) and (B) were isolated and analyzed by northern blot as described in Figure 2. Samples isolated from cells treated with UPF1 or UPF2 dsRNAs were analyzed in parallel. The numbers below the lanes indicate the levels of adh transcripts normalized to those of rp49 mRNA. (E) Normalized values obtained in three independent experiments for n4 were divided by those obtained for adh-wt. These ratios were set arbitrarily to 1 in control cells. (F) S2 cells expressing adh-n4 were transfected with dsRNA specific for UPF1, or with mixtures of dsRNAs as indicated. RNA samples were analyzed by northern blot. The numbers below the lanes indicate the levels of adh-n4 transcript normalized to those of rp49 mRNA.
Co-depletions of the EJC proteins severely impair cell proliferation (see Gatfield and Izaurralde, 2002). Nonetheless, no stabilization of adh-n4 mRNA was observed in cells depleted of single or multiple EJC proteins (Figure 6C–E). A major concern raised by the experiments described above was that the EJC proteins may have additional functions in mRNA metabolism that could mask the stabilization of adh-n4 mRNA. To address this issue, we simultaneously depleted Y14 or RNPS1 together with UPF1. We observed that in cells depleted of Y14 or RNPS1, depletion of UPF1 stabilized adh-n4 mRNA (Figure 6F). Similarly, depletion of RNPS1 and Y14 proteins together with Upf3 resulted in a stabilization of adh-n4 mRNA similar to that observed when only Upf3 was depleted (data not shown). We conclude that the EJC proteins are not essential for the degradation of adh-n4 mRNA.
PTCs can be defined independently of exon boundaries in Drosophila
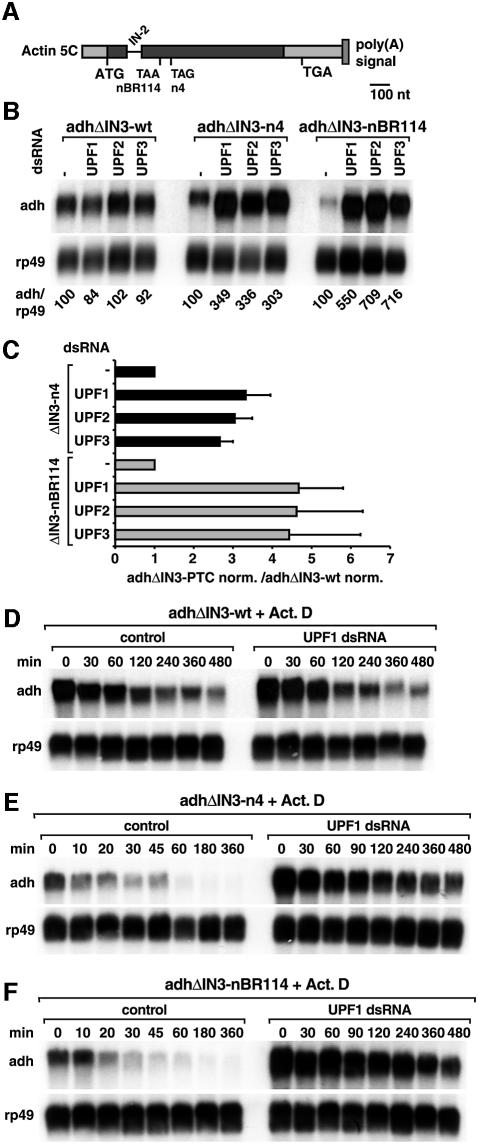
The observation that the expression levels of adh-n4 mRNA were not affected in cells depleted of EJC proteins raised the question as to whether exon–exon boundaries play a role in defining PTCs in Drosophila. To address this question, we deleted intron 3 (IN-3), which is the 3′-most intron in our reporters (Figure 7A), and generated cell lines expressing adhΔIN3-wt, adhΔIN3-n4 or adhΔIN3-nBR114 mRNAs.
Fig. 7. PTCs are defined independently of downstream exon boundaries in Drosophila cells. (A) Schematic representation of the reporters lacking a downstream intron. Symbols are as in Figure 1A. (B) S2 cells expressing adh-wt, adh-n4 or adh-nBR114 reporters lacking intron 3 (adhΔIN3-wt, adhΔIN3-n4, adhΔIN3-nBR114) were treated with UPF1, UPF2 or UPF3 dsRNAs. Total RNA samples were analyzed as described in Figure 2. (C) The steady-state levels of adhΔIN3-n4 and adhΔIN3-nBR114 mRNAs were quantitated in at least three independent experiments and normalized to those of rp49 mRNA. These normalized values (adhΔIN3-PTC norm.) were divided by those obtained for adhΔIN3-wt. The mean values ± SDs are shown. (D–F) S2 cells constitutively expressing adhΔIN3-wt, adhΔIN3-n4 or adhΔIN3-nBR114 were treated with UPF1 dsRNA. Seven days after addition of UPF1 dsRNA, treated or untreated cells (control) were incubated with actinomycin D (5 µg/ml) for the times indicated above the lanes. Total RNA samples were isolated and analyzed by northern blot.
Normalized to the levels of the endogenous rp49 mRNA, the levels of PTC-containing transcripts in which intron 3 had been deleted were reduced to 25–30% of the corresponding wild-type adhΔIN3 levels (Figure 7B). Moreover, while the half-life of adhΔIN3-wt was ∼90 min, the half-life of adhΔIN3-nBR114 mRNA was ∼15 min (control panels in Figure 7D–F).
To assess whether adhΔIN3-n4 or adhΔIN3-nBR114 were subjected to NMD, we suppressed the expression of UPF1, UPF2 or UPF3 genes in cells constitutively expressing these reporters. Silencing of either of the UPF genes resulted in a 3- to 5-fold increase of the steady-state levels of the PTC-containing mRNAs lacking intron 3. The expression levels of the wild-type mRNA were not significantly changed (Figure 7B). The stabilization of the PTC-containing mRNAs lacking intron 3 was therefore within the same range as that observed for the corresponding transcripts carrying intron 3 (Figure 7C versus 4B).
The increased steady-state levels of the PTC-containing mRNAs lacking intron 3 in cells depleted of UPF1 is due to a decrease of their turnover rate. Indeed, the half-life of adhΔIN3-n4 and of adhΔIN3-nBR114 mRNAs increased to ∼1 and 2 h, respectively, following UPF1 depletion (Figure 7E and F). The half-life of the corresponding wild-type transcript remained unchanged (Figure 7D). In summary, at least in the case of the adh mRNA, PTCs can be recognized by the Drosophila NMD machinery regardless of any downstream exon boundary.
The Drosophila NMD machinery discriminates PTCs from natural stops on heterologous mRNAs
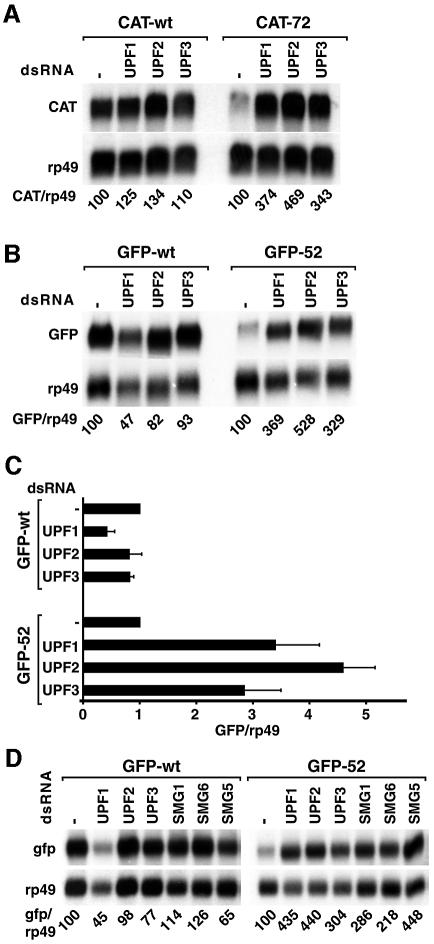
The exon boundary-independent recognition of PTCs described above could either be specific for the adh mRNA or could reflect a general feature of the NMD pathway in Drosophila. To discriminate between these possibilities, we generated two reporter genes in which either the CAT- (chloramphenicol acetyltransferase) or the GFP-coding regions were placed downstream of the constitutive actin 5C promoter. Both genes have no introns. The NMD reporters were generated by introducing a PTC at codon 72 and 52 of the CAT- and GFP-coding sequences, respectively. Polyclonal cell lines constitutively expressing wild-type or PTC-containing mRNAs were generated. Northern blot analysis showed that the levels of CAT-72 mRNA and GFP-52 mRNA were reduced in abundance to 30 and 15% of the corresponding wild-type levels (Figure 8A and B). For the GFP reporter, we confirmed that the reduction of the steady-state levels of GFP-52 mRNA correlated with an increase in its turnover rate. Indeed, the half-life of GFP-52 mRNA was ∼10 min, while the half-life of the wild-type GFP transcript was >4 h (data not shown).
Fig. 8. PTCs can be recognized on a heterologous mRNA. (A) S2 cells expressing CAT-wt or CAT-72 mRNAs were treated with UPF1, UPF2 or UPF3 dsRNAs. Total RNA samples were analyzed by northern blot using probes specific for CAT and rp49 mRNAs. The numbers below the lanes indicate the levels of the CAT transcripts normalized to those of rp49 mRNA. (B) S2 cells expressing GFP-wt or GFP-52 mRNAs were treated with UPF1, UPF2 or UPF3 dsRNAs. The levels of the GFP transcripts normalized to those of rp49 mRNA are indicated below the lanes. (C) The levels of GFP-wt and GFP-52 mRNAs were quantitated in at least three independent experiments and normalized to those of rp49 mRNA. These ratios were set arbitrarily to 1 in control cells. The mean values ± SDs are shown. (D) S2 cell lines expressing GFP-wt or GFP-52 were treated with the dsRNAs indicated above the lanes. Total RNA samples were analyzed as described in (B).
Depletion of either UPF1, UPF2 or UPF3 resulted in a 3- to 4.5-fold increase of the steady-state levels of the CAT-72 and GFP-52 mRNAs (Figure 8A–C), in agreement with the results obtained for the adh reporters. Surprisingly, a significant reduction in GFP-wt mRNA levels was consistently observed in UPF1-depleted cells (Figure 8B and C). Although the basis for this decrease is not known, it is possible that this effect reflects an additional function of UPF1 distinct from NMD, as GFP-wt mRNA levels were not significantly changed by UPF2 or UPF3 depletion (Figure 8B–D). Moreover, depletion of SMG1, SMG5 or SMG6 also led to a specific stabilization of GFP-52 mRNA (Figure 8D). We conclude that PTCs can be recognized by the Drosophila NMD machinery in heterologous mRNAs lacking a downstream exon boundary. These results provide strong support for the conclusion that the components of the EJC are dispensable for NMD in this organism.
Discussion
The NMD pathway in Drosophila provides a snapshot into the evolution of this conserved mRNA surveillance mechanism, as it has features in common with both the yeast and the mammalian pathways. In common with the yeast pathway is the observation that PTCs can be defined independently of exon boundaries. In contrast to the yeast pathway, however, NMD in Drosophila requires the products of the SMG1, SMG5 and SMG6 genes, which are a specific feature of higher eukaryotic NMD.
SMG5–7 proteins are a specific feature of the metazoan NMD pathway
Clissold and Ponting (2000) reported the existence of a protein family with N-terminal TPR repeats and a C-terminal PIN domain. Our multialignment of all sequences possessing these domains reveals that the conserved region detected by Clissold and Ponting (2000), which encompasses one and a half TPRs, can be extended to a region of ∼150 residues which is predicted to be helical over its entire length. We refer to this extended region as the TPR-like domain. TPR repeats are protein–protein interaction modules present in proteins with unrelated functions. As TPR motifs usually require more than one repeat for structural stability (Goebl et al., 1991), we predict that the region of extended similarity might contain several TPR repeats adapted to a specialized function in this protein family, so that standard computer programs may be unable to reveal statistically significant sequence similarities to known TPRs.
Previous studies identified three human proteins exhibiting limited sequence similarity to Ce SMG5 and SMG7 (Clissold and Ponting, 2000; Chiu et al., 2003; Reichenbach et al., 2003), but the orthologous relationships between these proteins had not been assigned. For that reason, Chiu et al. (2003) named KIAA0732, KIAA1089 and KIAA0250, human SMG5/7a, SMG5/7b and SMG5/7c, respectively. The nomenclature proposed by Chiu et al. (2003) is not supported by the phylogenetic trees based on the alignment of the TPR-like domain and of the PIN domain presented in this study. Indeed, the phylogenetic trees clearly define two different orthologous groups, the SMG5 family and the SMG6 family. As one gene of each organism falls into the same branch of the tree, they are likely to perform equivalent functions in these organisms, distinct from those of the second ortholog, but all involved in NMD. The nematode and human genomes encode in addition SMG7, but there is no obvious SMG7 ortholog in Drosophila. We therefore propose to call KIAA0732, KIAA1089 and KIAA0250, human SMG6, SMG5 and SMG7, respectively. Analogously, CG8954 and CG6369 will be named Drosophila SMG5 and SMG6, respectively. Given the distribution of proteins within the SMG5–7 families that have been shown to be involved in NMD (Cali et al., 1999; Page et al., 1999; Chiu et al., 2003; this study), we predict that all orthologs are likely to have a role in NMD, including those identified in Fugu rubripes and Ciona intestinalis (Figure 3).
Despite the similarites in domain organization, the SMG5–7 proteins do not appear to have redundant functions, as they are all required for the destabilization of PTC-containing transcripts. On the other hand, mutations in Ce smg-5, smg-6 or smg-7 genes lead to the accumulation of the phosphorylated form of UPF1 (Page et al., 1999), suggesting that all three proteins function in the dephosphorylation of UPF1. Consistently, Anders et al. (2003) have shown recently that Ce SMG5 is part of a protein complex including SMG7, UPF1, PP2Ac (the catalytic subunit of protein phosphatase 2A) and PR65 (a structural subunit of protein phosphatase 2A). Similarly, Chiu et al. (2003) have reported that human SMG6 co-immunoprecipitates with UPF1-3, SMG1 and PP2Ac. Together, these observations suggest that SMG5–7 are components of a conserved multiprotein complex, whose role is to direct PP2A to its UPF1 substrate. Depletion of any of the SMG5–7 proteins may lead to the disassembly of the complex and hence a similar phenotype, even though each protein probably has a distinct and specific function. Understanding the precise role of the SMG5–7 proteins in NMD awaits further biochemical characterization. In this context, the recently reported role of Hs SMG6 in telomere regulation (Reichenbach et al., 2003) suggests that some of these proteins have acquired additional functions distinct from NMD. Alternatively, since NMD regulates the expression levels of telomere components in yeast (Dahlseid et al., 2003), the possibility that SMG6 indirectly affects telomere maintenance in human cells cannot be ruled out.
Y14 and RNPS1 are dispensable for NMD in Drosophila
Human Y14:MAGOH and RNPS1 have been shown to elicit NMD when tethered downstream of a stop codon in human cells (Lykke-Andersen et al., 2001; Fribourg et al., 2003; Gehring et al., 2003). Moreover, depletion of Y14 from human cells by RNAi leads to the stabilization of PTC-containing transcripts, suggesting an essential role for Y14 in NMD (Gehring et al., 2003). Therefore, the observation that the Drosophila orthologs of vertebrate EJC proteins, and in particular Y14 and RNPS1, are dispensable for NMD reflects a specific feature of the Drosophila pathway. This observation is fully consistent with the observation that exon boundaries are not required to define PTCs in this organism.
Definition of premature stop codons in Drosophila
The observation that in the absence of a downstream exon boundary, PTC-containing mRNAs are nonetheless targeted for degradation raises the question of what differentiates a premature termination codon from a normal one in Drosophila. One possibility is that PTCs are defined relative to a DSE, as in yeast. Alternatively, a ‘fail-safe’ sequence may function instead of exon boundaries when the downstream intron is removed, as reported by Zhang et al. (1998a,b) for the β-globin and triosephosphate isomerase mRNAs. The observation that in-frame stop codons in the CAT or GFP mRNAs can be defined as premature by the Drosophila NMD machinery makes it unlikely that specific sequence elements mark Drosophila transcripts. However, because of the lack of a strong consensus among yeast DSEs or mammalian fail-safe sequences, we cannot rule out that the CAT or GFP mRNAs possess such sequence elements.
It is also possible that a generic feature of the mRNA, such as the poly(A) tail or a mark deposited during the cleavage and polyadenylation reaction, provides the positional information needed to discriminate premature from natural stop codons in Drosophila. The latter possibility is consistent with a model in which it is the environment of the translation termination codon that distinguishes a premature from a normal stop codon (Hilleren and Parker, 1999). According to this model, 3′-UTRs would be marked by a specific set of proteins that may become associated with the mRNA during 3′ end formation. If a terminating ribosome is able to interact with these 3′-UTR-bound proteins, proper termination can occur. If the termination process is impaired or too slow, because the terminating ribosome is unable to establish these interactions, the NMD complex could be assembled, leading to the rapid degradation of the mRNA.
Materials and methods
Drosophila NMD reporter constructs and cell lines
The adh sequence (accession No. X78384; residues 2021–2923) was amplified from a genomic clone and inserted between the EcoRI and XhoI sites of vector pAc5.1b (Invitrogen). The n4 and nBR114 mutations were inserted by site-directed mutagenesis using the Quick-change site-directed mutagenesis kit (Stratagene). Intron 3 was excised from the corresponding constructs by site-directed mutagenesis. A plasmid to allow the expression of GFP in S2 cells was generated by inserting the corresponding cDNA into a pBS-based vector containing the Drosophila actin promoter followed by multiple cloning sites and the BgH1 terminator (pBSactGFP). GFP-52 was derived from plasmid pBSactGFP by changing codon 52 (GGC) to a stop codon (TGA) in the GFP sequence. A plasmid to allow the expression of the CAT gene in S2 cells was generated by inserting the corresponding cDNA between the KpnI and XhoI sites of vector pAc5.1b. CAT-72 was generated by changing codon 72 (GAA) to a stop codon (TAA) in the CAT sequence. All constructs were sequenced completely to confirm the presence of the PTCs and the absence of additional mutations. Stable Drosophila cell lines expressing wild-type and PTC-containing transcripts were established by co-transfecting the corresponding constructs with a plasmid encoding puromycin acetyl transferase (pBS-PURO; Benting et al., 2000) at a 15:1 ratio. S2 cells were transfected with Lipofectin (Invitrogen) according to the manufacturer’s instructions and selected in medium containing 10 µg/ml puromycin.
Cloning of Drosophila cDNAs
Drosophila genes described in this study correspond to the following genes in FlyBase (http://flybase.bio.indiana.edu): UPF1 (CG1559), UPF2 (CG2253), UPF3 (CG11184), SMG1 (CG4549), SMG5 (CG8954, anon-34Ea) and SMG6 (CG6369). Full-length Drosophila UPF1 and UPF3 cDNAs were amplified by PCR using primers introducing unique restriction sites and a random-primed S2 cell cDNA as a template. For UPF2, SMG1, SMG5 and SMG6, cDNA fragments comprising only the first 700 nucleotides were cloned. All PCRs were performed with the Expand high-fidelity PCR system (Roche). The amplified cDNAs were cloned into pGEXCS and sequenced. Cloning of cDNAs encoding Drosophila orthologs of EJC proteins has been described before (Gatfield and Izaurralde, 2002).
Western blotting and double-stranded RNA interference in cultured Drosophila cells
Preparation of total cell extracts, western blotting and RNAi were performed essentially as described by Gatfield and Izaurralde (2002). DsRNAs corresponding to EJC proteins have also been described (Gatfield and Izaurralde, 2002). Other dsRNAs used in this study correspond to fragments comprising nucleotides 1–700 of the predicted cDNAs. A 30 µg aliquot of dsRNA was used per 6-well dish containing ∼2 × 106 cells. Depletions described in this study were carried out by transfecting cells with the corresponding dsRNAs on day 0 and day 4. Cells were harvested on day 7.
Double-stranded RNA interference in HeLa cells
HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 µg/ml streptomycin. Depletion of endogenous proteins by RNAi was carried out using siRNAs as described by Elbashir et al. (2001), except that cells were transfected with Lipofectamine Plus reagent (Invitrogen) following the manufacturer’s instructions. The following target sequences were used: UPF1, AAGATGCAGTTCCGCT CCATT; SMG5, AAGTCTTCCTGGACTGGCTTC; SMG6, AAGCC AGTGATACAGCGAATT; SMG7, AACAGCACAGTCTACAAGCCA; and control, AAGCTGCGGTACGCCAACAAC. The siRNAs were transfected at a final concentration of 70 nM. At 24 h post-transfection, cells were trypsinized and seeded in 6-well plates. After a further 24 h, the cells were transfected with siRNAs (as above) together with 0.5 µg of each β-globin reporter plasmid (β-globin-wt or β-globin-NS39) and 0.5 µg of pEGFPC1-NXF1 as a tranfection control. The β-globin reporters have been described by Thermann et al. (1998). Cells were harvested 48 h after the second transfection.
RNA isolation and northern blots
Total RNA was isolated using TRIzol reagent (Life Technologies), separated in denaturing formaldehyde agarose gels (5–20 µg/lane) and blotted onto positively charged nylon membranes (GeneScreen Plus, NEN Life Science). 32P-labeled probes were generated by random priming or linear PCR using standard methods. Hybridizations were carried out overnight at 65°C in Church buffer (0.5 M Na-phosphate pH 7; 7% SDS and 1 mM EDTA). Following hybridization, the membranes were washed four times 15 min at 65°C in 40 mM Na-phosphate pH 7, 1% SDS and 1 mM EDTA.
Sequence analysis
Sequence similarity searches were performed using PSI-BLAST (Altschul et al., 1997) with Ce SMG-7 and SMG-5 as input sequences. These searches did not converge due to the presence of widespread domains such as TPR and PIN. Nevertheless, a group of putatively related sequences could be defined on the basis of the presence of both domains. The relationship between these proteins was confirmed by using each of the sequences as an input for PSI-BLAST and retrieving all the other members of the group. The human sequence KIAA0250 (BAC53621) was joined to the sequences bearing the two domains, as it was retrieved in all the searches. As the identified proteins show no sequence conservation outside of the TPR-like and PIN domains, two different multialignments and the corresponding bootstrap trees were built with ClustalW (Thompson et al., 1997) using only these domains. Because of the low degree of conservation of the extended TPR-like region, the bootstrap values for this tree were not as good as those obtained for the PIN domain.
Acknowledgments
Acknowledgements
We gratefully acknowledge Saverio Brogna for providing a genomic adh clone, Philip Anderson for providing information on the identity of C.elegans SMG6, Philipp Bucher for providing unpublished results on the alignment of the SMG5-7 proteins, Kevin Czaplinski and Matthias Hentze for helpful and stimulating discussions, and David Thomas for critical reading of the manuscript. This study was supported by the European Molecular Biology Organization (EMBO).
References
- Adams M.D. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- Anders K.R., Grimson,A. and Anderson,P. (2003) SMG-5, required for C.elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J., 22, 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benting J., Lecat,S., Zacchetti,D. and Simons,K. (2000) Protein expression in Drosophila Schneider cells. Anal. Biochem., 278, 59–68. [DOI] [PubMed] [Google Scholar]
- Birney E., Thompson,J.D. and Gibson,T.J. (1996) PairWise and SearchWise: finding the optimal alignment in a simultaneous comparison of a protein profile against all DNA translation frames. Nucleic Acids Res., 24, 2730–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogna S. (1999) Nonsense mutations in the alcohol dehydrogenase gene of Drosophila melanogaster correlate with an abnormal 3′ end processing of the corresponding pre-mRNA. RNA, 5, 562–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cali B.M., Kuchma,S.L., Latham,J. and Anderson,P. (1999) smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics, 151, 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S.Y., Serin,G., Ohara,O. and Maquat,L.E. (2003) Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA, 9, 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clissold P.M. and Ponting,C.P. (2000) PIN domains in nonsense-mediated mRNA decay and RNAi. Curr. Biol., 10, R888–R890. [DOI] [PubMed] [Google Scholar]
- Dahlseid J.N. et al. (2003) mRNAs encoding telomerase components and regulators are controlled by UPF genes in Saccharomyces cerevisiae. Eukaryot. Cell, 2, 134–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning G., Jamieson,L., Maquat,L.E., Thompson,E.A. and Fields,A.P. (2001) Cloning of a novel phosphatidylinositol kinase-related kinase: characterization of the human SMG-1 RNA surveillance protein. J. Biol. Chem., 276, 22709–22714. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Fribourg S., Gatfield,D., Izaurralde,E. and Conti,E. (2003) A novel mode of RBD-protein recognition in the Y14–Mago complex. Nat. Struct. Biol., 10, 433–439. [DOI] [PubMed] [Google Scholar]
- Gatfield D. and Izaurralde,E. (2002) REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J. Cell Biol., 159, 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring N.H., Neu-Yilik,G., Schell,T., Hentze,M.W. and Kulozik,A.E. (2003) Y14 and hUpf3b form an NMD-activating complex. Mol. Cell, 11, 939–949. [DOI] [PubMed] [Google Scholar]
- Goebl M. and Yanagida,M. (1991) The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem. Sci., 16, 173–177. [DOI] [PubMed] [Google Scholar]
- He F., Brown,A.H. and Jacobson,A. (1997) Upf1p, Nmd2p and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol., 17, 1580–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P. and Parker,R. (1999) mRNA surveillance in eukaryotes: kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA, 5, 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N., Yong,J., Kataoka,N., Abel,L., Diem,M.D. and Dreyfuss,G. (2001a) The Y14 protein communicates to the cytoplasm the position of exon–exon junctions. EMBO J., 20, 2062–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N., Kataoka,N. and Dreyfuss,G. (2001b) Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon–exon junction complex. Science, 293, 1832–1836. [DOI] [PubMed] [Google Scholar]
- Le Hir H., Gatfield,D., Izaurralde,E. and Moore,M.J. (2001) The exon–exon junction complex provides a binding platform for factors involved in mRNA export and NMD. EMBO J., 20, 4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu,M.-D. and Steitz,J.A. (2000) Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell, 103, 1121–1131. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu,M.D. and Steitz,J.A. (2001) Communication of the position of exon–exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science, 293, 1836–1839. [DOI] [PubMed] [Google Scholar]
- Mendell J.T., ap Rhys,C.M.J. and Dietz,H.C. (2002) Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science, 298, 419–422. [DOI] [PubMed] [Google Scholar]
- Nagy E. and Maquat,L.E. (1998) A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci., 23, 198–199. [DOI] [PubMed] [Google Scholar]
- Page M.F., Carr,B., Anders,K.R., Grimson,A. and Anderson,P. (1999) SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol., 19, 5943–5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M., Ishigaki,Y., Nagy,E. and Maquat,L.E. (2001) Evidence that phosphorylation of human Upf1 protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA, 7, 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach P., Hoss,M., Azzalin, C.M., Nabholz,M., Bucher,P. and Lingner,J. (2003) A human homolog of yeast est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr. Biol., 13, 568–574. [DOI] [PubMed] [Google Scholar]
- Serin G., Gersappe,A., Black,J.D., Aronoff,R. and Maquat,L.E. (2001) Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol. Cell. Biol., 21, 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann R., Neu-Yilik,G., Deters,A., Frede,U., Wehr,K., Hagemeier,C., Hentze,M.W. and Kulozik,A.E. (1998) Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J., 17, 3484–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. and Lykke-Andersen,J. (2002) mRNA surveillance: the perfect persist. J. Cell Sci., 115, 3033–3038. [DOI] [PubMed] [Google Scholar]
- Yamashita A., Ohnishi,T., Kashima,I., Taya,Y. and Ohno,S. (2001) Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev., 15, 2215–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Sun,X., Qian,Y. and Maquat,L.E. (1998a) Intron function in the nonsense-mediated decay of β-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA, 4, 801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Sun,X., Qian,Y., LaDuca,J.P. and Maquat,L.E. (1998b) At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol. Cell. Biol., 18, 5272–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]