Abstract
Group II introns are ribozymes whose catalytic mechanism closely resembles that of the spliceosome. Many group II introns have lost the ability to splice autonomously as the result of an evolutionary process in which the loss of self-splicing activity was compensated by the recruitment of host-encoded protein cofactors. Genetic screens previously identified CRS1 and CRS2 as host-encoded proteins required for the splicing of group II introns in maize chloroplasts. Here, we describe two additional host-encoded group II intron splicing factors, CRS2-associated factors 1 and 2 (CAF1 and CAF2). We show that CRS2 functions in the context of intron ribonucleoprotein particles that include either CAF1 or CAF2, and that CRS2–CAF1 and CRS2–CAF2 complexes have distinct intron specificities. CAF1, CAF2 and the previously described group II intron splicing factor CRS1 are characterized by similar repeated domains, which we name here the CRM (chloroplast RNA splicing and ribosome maturation) domains. We propose that the CRM domain is an ancient RNA-binding module that has diversified to mediate specific interactions with various highly structured RNAs.
Keywords: chloroplast/CRM domain/group II intron/UPF 0044
Introduction
Group II introns are large, catalytic RNAs that are defined by limited regions of conserved sequence, a conserved structural organization consisting of six largely helical domains and characteristic inter-domain interactions (Michel et al., 1989; Michel and Ferat, 1995; Qin and Pyle, 1998; Bonen and Vogel, 2001). They are widely distributed in eubacteria, mitochondria and chloroplasts, and sporadically represented in the archaea (Lambowitz et al., 1999; Dai and Zimmerly, 2002, 2003; Toro, 2003). There is strong evidence that group II introns share a common ancestor with nuclear pre-mRNA introns and the spliceosome (Cech, 1986; Sharp, 1991; Michel and Ferat, 1995; Nilsen, 1998; Collins and Guthrie, 2000; Villa et al., 2002). Support for this hypothesis comes from the overall similarity of their splicing chemistry, similar RNA structures in their catalytic cores, experiments suggesting spliceosomal RNA is catalytic (Valadkhan and Manley, 2001) and evidence for catalytic Mg2+ ions bound to RNA moieties in both instances (Sontheimer et al., 1997; Gordon et al., 2000; Gordon and Piccirilli, 2001; Villa et al., 2002).
Many group II introns are mobile genetic elements. Their mobility is mediated by a ribonucleoprotein (RNP) complex composed of intron RNA and a conserved intron-encoded protein (IEP), often called a maturase, that also facilitates the splicing of its host intron (Eickbush, 1999; Lambowitz et al., 1999). It is believed that mobile group II introns were introduced into primordial eukaryotic cells through the incorporation of bacterial endosymbionts, permitting these introns to invade the nuclear genome. Subsequent degeneration of the introns may then have been accompanied by the recruitment of host proteins to form a trans-acting splicing complex and, ultimately, the spliceosome. Properties of contemporary group II introns are consistent with the idea that group II introns co-evolve with their hosts to produce novel splicing RNPs. Most organellar group II introns lack genes for IEPs, although they appear to have evolved from IEP-encoding ancestors (Toor et al., 2001). IEP loss seems to have been accompanied by the recruitment of host-encoded proteins to facilitate splicing, yielding splicing-competent RNPs harboring degenerate group II introns that have lost the capacity for autocatalytic splicing. The requirement for host-encoded proteins in splicing permits the incorporation of regulated splicing into the biology of the host organism (Lambowitz et al., 1999).
The chloroplasts of higher plants provide a useful venue for the study of group II intron–host factor co-evolution because they have many group II introns, most of which do not encode an IEP, and none of which have been observed to self-splice in vitro. Thus, these introns are likely to require host-encoded proteins for efficient splicing in vivo. Indeed, genetic screens have identified host-encoded proteins required for the splicing of 10 of the 17 group II introns in the maize chloroplast (Jenkins et al., 1997; Jenkins and Barkan, 2001; Till et al., 2001). Analogous screens in the unicellular chlorophyte Chlamydomonas reinhardtii have shown that the splicing of its two chloroplast group II introns also involves the participation of host-encoded proteins (Goldschmidt-Clermont et al., 1990; Perron et al., 1999; Rivier et al., 2001).
The maize proteins chloroplast RNA splicing 1 (CRS1) and 2 (CRS2) are required for the splicing of different subsets of the group II introns in the chloroplast (Jenkins et al., 1997). CRS1, which is required solely for the splicing of the atpF pre-mRNA, is the founding member of a family of plant proteins containing a novel RNA-binding domain of ancient origin (Till et al., 2001; Ostheimer et al., 2002; Willis et al., 2002). CRS2 is required for the splicing of nine of the 10 chloroplast introns in subgroup IIB (Jenkins et al., 1997), which is one of two group II intron subgroups that are distinguished by subtle structural differences (Michel et al., 1989). CRS2 is closely related to bacterial peptidyl-tRNA hydrolase (PTH) (Jenkins and Barkan, 2001), an enzyme that hydrolyzes the ester bond linking the tRNA and nascent polypeptide in abortive translation products (Menninger, 1976).
In this work, we show that CRS2 forms complexes with two other proteins, CRS2-associated factors 1 and 2 (CAF1 and CAF2). The CAFs are, themselves, splicing factors, required for the splicing of different subsets of the CRS2-dependent introns. Furthermore, the CRS2–CAF1 and CRS2–CAF2 complexes are bound in vivo to their target group II intron RNAs. These and other findings provide strong evidence that CRS2 forms functional splicing complexes with either CAF1 or CAF2 in vivo, and that the CAF subunit determines the intron specificity of the complex.
Interestingly, CAF1 and CAF2 are members of a protein family in maize that includes the previously identified group II intron splicing factor CRS1. The similarity between these proteins is confined to repeated segments corresponding to the novel RNA-binding domain initially identified in CRS1 (Till et al., 2001; Ostheimer et al., 2002; Willis et al., 2002). Thus, as the group II introns of higher plant chloroplasts co-evolved with the nuclear genome, they appear to have spurred the evolution of a family of intron-specific group II intron splicing factors by amplification and diversification of an ancient RNA-binding module.
Results
Discovery of CAFs in a yeast two-hybrid screen
CRS2 is related to bacterial PTH, which is a monomeric protein (Schmitt et al., 1997). However, several observations suggested that CRS2 promotes splicing as a component of a protein complex. First, CRS2 was not sufficient to promote splicing of its cognate introns in vitro, or when co-expressed with plastid pre-mRNAs in Escherichia coli (data not shown). Secondly, CRS2, a protein of 23 kDa, is found in a nuclease-resistant complex of ∼80 kDa in chloroplast extract (Jenkins and Barkan, 2001). Finally, the crystal structure of CRS2 (G.J.Ostheimer, A.Barkan and B.W.Matthews, in preparation) demonstrated that CRS2 possesses a surface-exposed hydrophobic patch suggestive of a protein interaction surface.
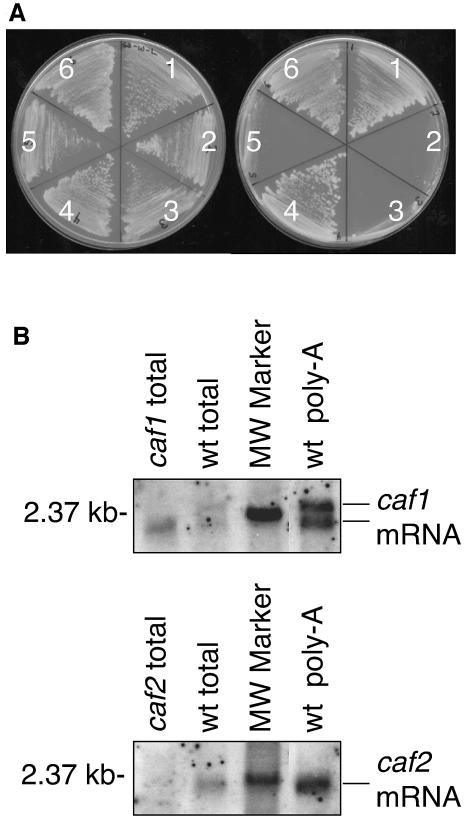
A yeast two-hybrid screen (Fields and Song, 1989) was used to identify maize proteins that can interact with CRS2. The bait plasmid encoded CRS2 fused to the Gal4 DNA-binding domain. The library of prey plasmids consisted of a maize leaf cDNA library cloned into a Gal4 activation domain fusion vector. Among 7 × 105 co-transformants, six were identified that survived selection for growth in the absence of histidine (Figure 1A) and also tested positive in a secondary β-galactosidase screen (not shown). The six positive clones were derived from two uncharacterized but closely related genes, which we named caf1 and caf2. Two of the cDNAs originated from the caf1 gene, but had distinct 5′ ends and polyadenylation sites. Four of the cDNAs originated from the caf2 gene, three of which were identical in sequence and the fourth differing in the position of its 5′ end.
Fig. 1. Identification of CAF1 and CAF2. (A) CRS2 interacts with CAF1 and CAF2 in a yeast two-hybrid assay. Yeast strains were streaked on plates containing (left) and lacking histidine (right). Growth in the absence of histidine indicates a positive two-hybrid interaction. Cells contained plasmids encoding fusion proteins to the Gal4 DNA-binding domain (BD) and transcription activation domain (AD), as follows: (1) BD and AD fused to a λcI fragment that homodimerizes (positive interaction control); (2) BD–CRS2 and AD–cI (negative control); (3) BD–cI and AD–CAF1 (negative control); (4) BD–CRS2 and AD–CAF1; (5) BD–cI and AD–CAF2 (negative control); and (6) BD–CRS2 and AD–CAF2. (B) RNA gel blot hybridization showing the size of the caf1 and caf2 mRNAs. Lanes contain 0.5 µg of poly(A)-enriched RNA from wild-type seedling leaf (wt poly-A), or 20 µg of total seedling leaf RNA from wild-type, caf1 mutant or caf2 mutant. The upper panel was probed with a caf1 gene-specific probe; the lower panel was probed with a caf2 gene-specific probe. The position of a 2.37 kb RNA marker is indicated.
RNA gel blot hybridization of maize leaf RNA revealed a single caf2 mRNA of ∼2.3 kb, and two caf1 mRNAs, of ∼2.3 and ∼2.4 kb (Figure 1B). Alternative caf1 polyadenylation sites separated by 180 nucleotides were revealed by the caf1 cDNA sequences (not shown), and could account for the caf1 mRNA doublet. The longest cDNAs recovered [excluding poly(A) sequences] were 2312 and 2200 bp for caf1 and caf2, respectively. These cDNAs encode long open reading frames (ORFs; 674 amino acids for caf1 and 611 amino acids caf2), beginning with start codons near their 5′ ends. Taken together, these results suggest that the ORFs are full length and that the intact mRNAs include an additional ∼100 nucleotides of 5′-untranslated region (UTR) and poly(A) tail sequences.
Database homology searches identified a family of predicted plant proteins similar to CAF1 and CAF2. Putative orthologs were identified as proteins with higher levels of similarity to CAF1 or CAF2 than to other members of the CAF family within the same species. An alignment of maize CAF1 and CAF2 with putative rice and Arabidopsis orthologs is shown in Figure 2. All of these proteins are predicted to be targeted to the chloroplast (http://www.inra.fr/Internet/Produits/Predotar; Emanuelsson et al., 2000) consistent with their functioning in complex with CRS2.
Fig. 2. Alignment of CAF1 and CAF2 with predicted orthologs in rice and Arabidopsis. Identical residues are shaded in black and similar residues in gray. The predicted cleavage sites of the chloroplast targeting sequences are indicated by diamonds. Double arrowheads show the starting points of truncated CAFs identified in the yeast two-hybrid screen. The positions of the Mu insertions in the caf mutants analyzed in Figure 5 are shown by inverted triangles. Alignments were calculated with ClustalW (Higgins et al., 1994) and shaded with BOXSHADE. Accession Nos: rice CAF1, BAC05662; rice CAF2, BAB21243; Arabidopsis CAF1, At2g20020; Arabidopsis CAF2, At1g23400. The CAF1 and CAF2 sequences have been deposited in GenBank under accession Nos AY264368 and AY264369, respectively.
CAF1 and CAF2 are related to the chloroplast group II intron splicing factor CRS1
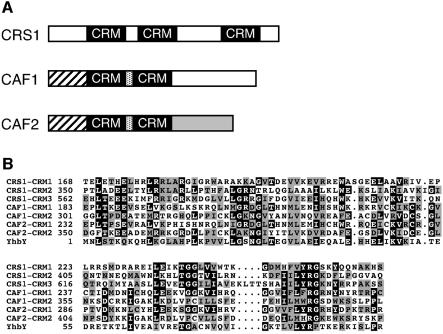
Database searches also revealed a set of more distant CAF1 and CAF2 relatives in plants that, intriguingly, included CRS1, a previously identified host-encoded group II intron splicing factor (Jenkins et al., 1997; Till et al., 2001). CRS1 is required for the splicing of the group IIA intron in the maize chloroplast atpF gene (Jenkins et al., 1997). CRS1 was noted previously to include three copies of a domain of ancient origin, represented as a free-standing ORF in eubacteria and archaea (Till et al., 2001). The similarity between CRS1 and the CAFs is confined to these domains, which occur twice in each of the CAFs (Figures 2 and 3).
Fig. 3. CRM domains in group II intron splicing factors. (A) Schematic illustrating regions of similarity between CRS1, CAF1 and CAF2. Similar shading indicates amino acid sequence similarity. (B) Alignment of CRM domains in CAF1, CAF2 and CRS1 with E.coli YhbY. Residue numbers indicate the position of each CRM domain within the context of the full-length protein. The complete YhbY sequence is shown.
The E.coli protein YhbY is typical of the predicted prokaryotic proteins that align with the repeated domains in CRS1, CAF1 and CAF2. We have shown recently that YhbY is bound tightly and specifically to pre-50S ribosomal subunits, suggesting that it facilitates ribosome maturation (T.Kawamura and A.Barkan, in preparation). Therefore, we propose that the plant domains and their prokaryotic homologs be called chloroplast RNA splicing and ribosome maturation, or CRM, domains. An alignment of the maize splicing factor CRM domains and E.coli YhbY is shown in Figure 3B. The crystal structure of YhbY provided evidence that the CRM domain constitutes a previously unrecognized RNA-binding domain (Ostheimer et al., 2002; Willis et al., 2002). Their possession of CRM domains and their interaction with CRS2 in a yeast two-hybrid assay strongly suggested that the CAFs are themselves group II intron splicing factors, and that they interact directly with both CRS2 and intron RNA.
CRS2 is bound to CAF1 and CAF2 in chloroplasts
To test whether CRS2 associates with CAF1 and CAF2 in vivo, antisera raised to CAF1- or CAF2-specific antigens were used in co-immunoprecipitation experiments with chloroplast stroma. Both antisera efficiently co-immunoprecipitated CRS2, whereas other antisera did not (Figure 4). Therefore, CAF1 and CAF2 are bound to CRS2 in chloroplasts, and the yeast two-hybrid interactions reflect bona fide in vivo interactions. The CAF1 antiserum did not detectably co-precipitate CAF2, and vice versa (Figure 4), suggesting that CRS2 interacts with either CAF1 or CAF2, but not with both simultaneously.
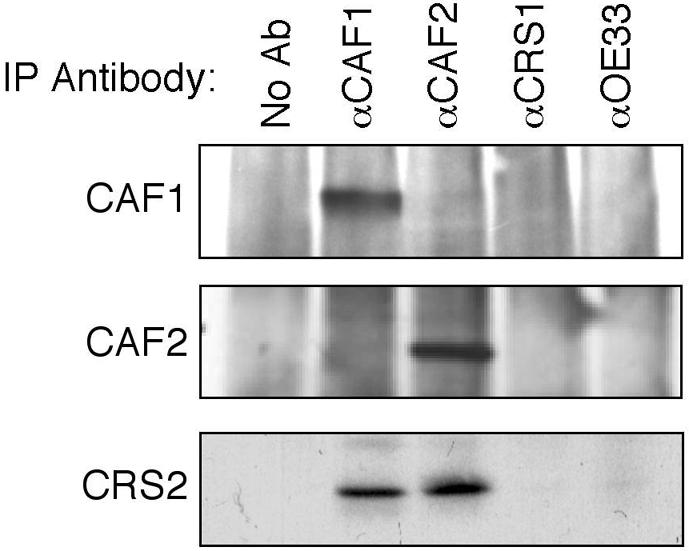
Fig. 4. Co-immunoprecipitation of CRS2 with CAF1 and CAF2 from chloroplast extract. Chloroplast stroma was incubated with the affinity-purified antiserum indicated above each lane. Immunoprecipitates were analyzed on immunoblots to detect CAF1 (top panel), CAF2 (middle panel) or CRS2 (bottom panel). Antiserum to OE33, which is a component of the photosynthetic apparatus that does not bind RNA, was used as a negative control. The αCRS1 immunoprecipitation successfully precipitated CRS1 (not shown) and the CRS1-dependent atpF intron (see Figure 6).
CAF1 and CAF2 are required in vivo for the splicing of chloroplast group II introns
The similarity of the CAFs to the group II intron splicing factor CRS1 and their physical association with the splicing factor CRS2 strongly suggested that these proteins likewise function in group II intron splicing. To test this idea, caf mutants were obtained through a reverse genetic screen. Pooled DNA samples from our collection of ∼2000 Mu transposon-induced, non-photosynthetic maize mutants (http://chloroplast.uoregon.edu) were screened by PCR with caf gene-specific primers in conjunction with a Mu primer. A DNA fragment is amplified when a Mu element is inserted near sequences bound by the gene primer. One mutant caf2 allele (caf2-1) and two mutant caf1 alleles (caf1-1 and caf1-2) were recovered; the positions of the Mu insertions in these alleles are illustrated on the alignment in Figure 2. The mutants have the same ‘ivory’ leaf phenotype as crs2 mutants, consistent with disruptions in the same pathway. caf2-1 is likely to be a null allele because its Mu insertion is well within the ORF (Figure 2). The caf1 alleles may be leaky, however, because their Mu insertions disrupt sequences near the beginning of the ORF (Figure 2), and northern blots of mutant RNA detected some caf1 mRNA of near normal size (Figure 1B). Transcription proceeding outward from the ends of the Mu transposons (Barkan and Martienssen, 1991) may be responsible for the aberrant caf1 mRNA.
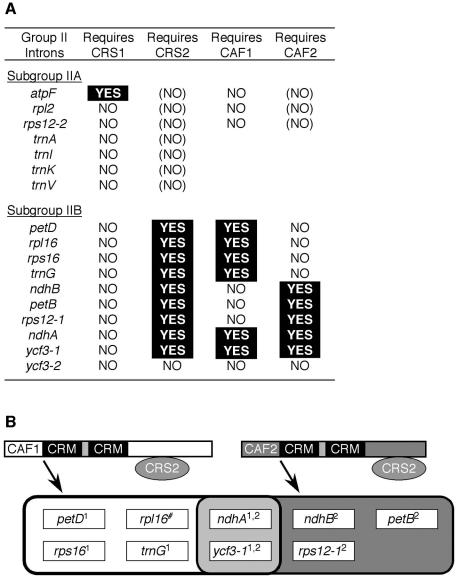
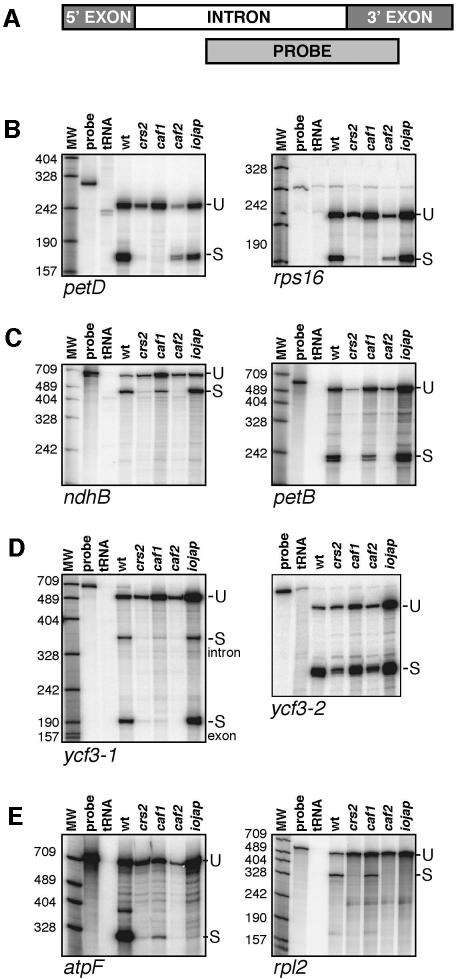
The maize chloroplast genome has 17 group II introns including members of subgroups IIA and IIB, as summarized in Figure 7. The splicing of 13 of these introns in homozygous caf1 and caf2 mutants was analyzed with RNase protection assays. Fully albino iojap mutants were analyzed in parallel. These mutants lack plastid ribosomes and illustrate the subgroup IIA-specific splicing defects that result from severe plastid ribosome deficiencies (Jenkins et al., 1997; Vogel et al., 1999). Representative RNase protection data are shown in Figure 5, and the full data set is summarized in Figure 7. The results show that the CAFs are, indeed, required for the splicing of chloroplast group II introns in vivo. Furthermore, all introns that require CRS2 also require a CAF, and the one CRS2-independent subgroup IIB intron (ycf3-2) also splices independently of CAF1 and CAF2. This correlation suggests that CRS2 and the CAFs always facilitate splicing in conjunction with one another. Strikingly, the nine CRS2-dependent introns can be subdivided according to their requirement for CAF1 or CAF2 (Figure 7). Four introns (petD, rpl16, rps16 and trnG) require CAF1 but not CAF2, three introns (ndhB, petB and rps12-1) require CAF2 but not CAF1, and two introns (ndhA and ycf3-1) require both CAF1 and CAF2.
Fig. 7. Summary of intron targets of four host-encoded group II intron splicing factors in maize chloroplasts. (A) The maize chloroplast group II introns and their requirements for CRS1, CRS2, CAF1 and CAF2. Requirements were deduced from the splicing defects detected in crs1, crs2, caf1 and caf2 mutants. The crs1 and crs2 mutant data are from Jenkins et al. (1997) and Vogel et al. (1999). The caf mutant data are from this work. Although crs2-1 and caf2-1 mutants exhibit defects in subgroup IIA splicing, these are thought to be indirect effects of their plastid ribosome deficiency and are therefore designated as (NO) in this table. (B) Diagram summarizing both the genetic and biochemical intron specificities of CRS2–CAF1 and CRS2–CAF2 complexes. Arrows point to the set of introns that require either CAF1 or CAF2 to splice in vivo. Superscripts indicate whether CAF1 antiserum (superscript 1) or CAF2 antiserum (superscript 2) strongly co-immunoprecipitated the indicated intron. The hash sign indicates that the rpl16 intron was poorly precipitated by both antisera.
Fig. 5. RNase protection analysis of group II intron splicing defects in caf1 and caf2 mutants. The RNase protection probes used here have been described previously (Jenkins et al., 1997). They spanned either the 5′- or the 3′-splice junction. Total leaf RNA from wild-type (wt) or the indicated mutant line was analyzed. The crs2-1 allele analyzed here conditions defects in both subgroup IIA and subgroup IIB splicing (Jenkins et al., 1997). tRNA indicates reactions in which tRNA was substituted for leaf RNA. Unspliced RNA protects both intron and exon probe sequence (U), whereas spliced RNA protects exon sequence (S) and, in some cases, excised intron sequence (S-intron). (A) Schematic of a 3′-splice site probe. (B) Representative CAF1-dependent introns. (C) Representative CAF2-dependent introns. (D) The ycf3 introns. ycf3-1 requires both CAF1 and CAF2 for splicing. The ycf3-2 intron, which is the only subgroup IIB intron that does not require CRS2, likewise does not require CAF1 or CAF2. (E) Representative subgroup IIA introns. These introns are not spliced in mutants such as iojap, with severe plastid ribosome deficiencies. Evidence presented in Jenkins et al. (1997) suggested that the subgroup IIA splicing defect in the crs2-1 allele is a pleiotropic effect of its plastid ribosome loss. Intron co-immunoprecipitation data presented in Figure 6 suggest the same is true for caf2 mutants.
In addition to defects in the splicing of several CRS2-dependent subgroup IIB introns, the caf2 mutants also failed to splice subgroup IIA introns (Figure 5E). The interpretation of these subgroup IIA splicing defects is complicated by the fact that subgroup IIA introns remain unspliced in mutants lacking plastid ribosomes (see iojap in Figure 5E) (Jenkins et al., 1997; Vogel et al., 1999). Therefore, the subgroup IIA defects in the caf2 mutants could either reflect a direct role for CAF2 in subgroup IIA splicing, or could be a secondary effect of a plastid ribosome deficiency. Previously we found that the null crs2-1 allele (the allele assayed in Figure 5) also conditions defects in both group IIA and IIB splicing, whereas the ‘leaky’ crs2-2 allele conditions defects solely in group IIB splicing (Jenkins et al., 1997). This led to the conclusion that CRS2 functions directly in subgroup IIB splicing, with the subgroup IIA defects in the null allele resulting from the failure to splice RNAs involved in translation (i.e. rps16, rpl16, trnG and rps12), and the consequent loss of plastid ribosomes. The co-immunoprecipitation data described below lead us to favor the idea that the subgroup IIA splicing defects in the caf2 mutant are likewise a pleiotropic effect due to its lack of plastid ribosomes, and do not reflect a direct role for CAF2 in their splicing.
CAF1 and CAF2 are found in complex with their genetically defined intron targets in vivo
Figure 4 showed that αCAF1 and αCAF2 antibodies co-immunoprecipitate CRS2 from chloroplast extract. To determine whether intron RNAs are bound tightly by the CRS2–CAF complexes in vivo, RNA was extracted from immunoprecipitate pellets and supernatants and analyzed by slot-blot hybridization, using probes to various chloroplast introns. To provide a point of comparison, αCRS1 immunoprecipitations were performed in parallel: CRS1 is also a CRM domain splicing factor, but it differs from the CAFs in that it does not work in concert with CRS2 and is required solely for the splicing of the atpF intron (Jenkins et al., 1997). The CRS1 antibody efficiently co-precipitated atpF intron RNA but did not precipitate any of the CRS2–CAF-dependent introns (Figure 6; data not shown). Analogous results were obtained with the αCAF1 and αCAF2 sera: the CAF1-dependent introns trnG, petD and rps16 were strongly enriched in the αCAF1 immunoprecipitates, but not in the αCAF2 or αCRS1 precipitates (Figure 6; data not shown). The CAF2-dependent introns ndhB, petB and rps12-1 were strongly enriched in the αCAF2 precipitates but not in the αCAF1 or αCRS1 precipitates (Figure 6; data not shown). The ycf3-1 and ndhA introns, which are unusual in requiring both CAF1 and CAF2 for their splicing, were co-immunoprecipitated by both αCAF1 and αCAF2 sera (Figure 6; data not shown). These results show that CRS1 and the CRS2–CAF complexes are bound tightly and preferentially to their genetically defined group II intron targets in vivo.
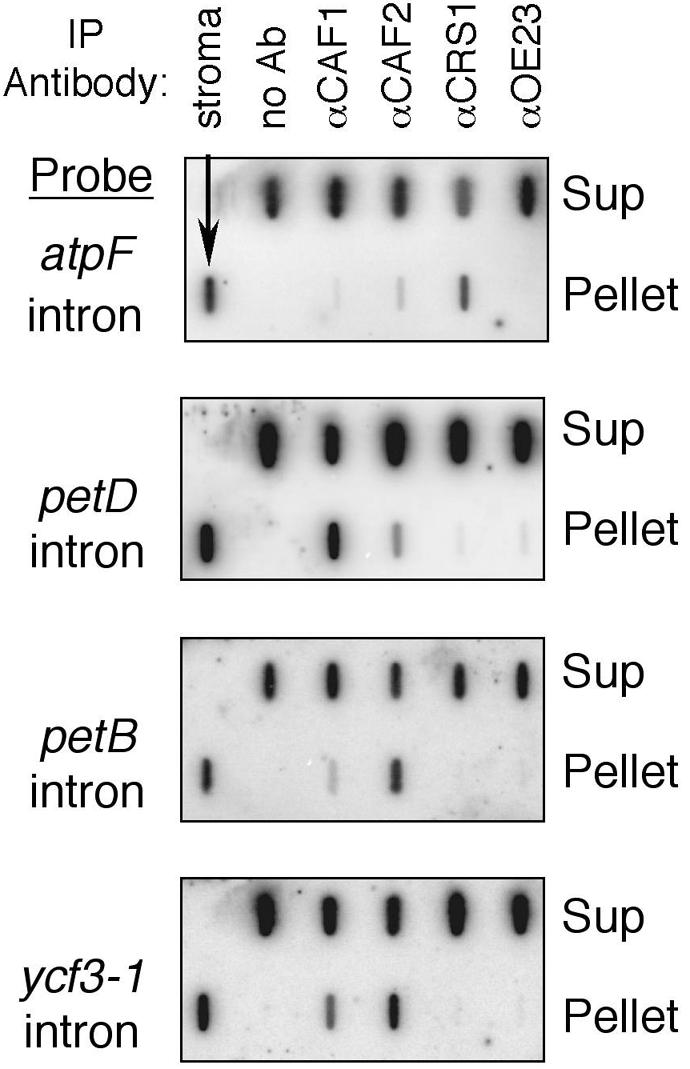
Fig. 6. Co-immunoprecipitation of group II intron RNA with CAF1 and CAF2 from chloroplast extract. Chloroplast stroma was subject to immunoprecipitation with the indicated affinity-purified antisera. RNA was extracted from the immunoprecipitate pellet and supernatant (Sup) and applied to a nylon membrane with a slot-blot manifold. Duplicate slot-blots were hybridized with probes specific for the indicated intron. Negative controls included a mock precipitation (no Ab), and precipitation with antisera to OE23, which is a component of the photosynthetic apparatus that does not bind RNA. RNA extracted from total stroma was applied to one slot on each blot as a positive hybridization control.
There is good correspondence between the set of introns that is co-precipitated by each antibody and the set whose splicing is disrupted in the corresponding mutant background. However, the co-immunoprecipitation data suggest that CAF1 and CAF2 bind introns more promiscuously than is indicated by the mutant phenotypes. For example, αCAF2 inefficiently but detectably co-precipitates the CAF1-dependent petD intron, but CAF2 is not required for splicing of the petD intron (Figure 5). Analogously, the αCAF1 serum weakly precipitates the CAF2-dependent ndhB intron, and CAF1 is not required for ndhB intron splicing (not shown). Given the extensive sequence identity between CAF1 and CAF2 (Figure 2), it would not be surprising if the RNA binding specificities of the CRS2–CAF1 and CRS2–CAF2 complexes overlap. Whereas the genetic data support the idea that the major CRS2–CAF–intron complexes detected in the co-immunoprecipitations reflect interactions along the productive splicing pathway, the genetic data also show that the minor complexes are not necessary for efficient splicing in vivo. These minor complexes may represent non-productive interactions, or interactions that contribute to the splicing of only a small proportion of intron molecules. Two of the nine CRS2-dependent introns, ndhA and ycf3-2, fail to splice in either caf1 or caf2 mutants and are precipitated from wild-type extract by both αCAF antisera (Figure 7), suggesting that these introns form splicing RNPs that contain both CRS2–CAF1 and CRS2–CAF2 complexes.
As discussed above, it was unclear whether the subgroup IIA splicing defects in the caf2 mutants were a secondary effect of the ribosome deficiency in caf2 mutant plastids. The co-immunoprecipitation data help to clarify this issue. The αCAF2 antiserum efficiently co-precipitated the five subgroup IIB introns whose splicing requires CAF2 (ndhB, ndhA, rps12-1, petB and ycf3-1) but not the subgroup IIA atpF or rpl2 introns (Figures 6 and 7). These data support the idea that the subgroup IIA splicing defects in the caf2 mutants are a secondary effect, and do not reflect a direct role for CAF2 in the splicing of the subgroup IIA introns.
Discussion
In this work, we describe the discovery of CAF1 and CAF2, two closely related proteins that function in concert with CRS2 to promote the splicing of group II introns in maize chloroplasts. CRS2 interacts with CAF1 and CAF2 in a yeast two-hybrid assay (Figure 1) and after co-expression in E.coli (not shown), indicating that the interactions are direct. The CRS2–CAF interactions are functionally relevant, as caf1 and caf2 mutants are defective in the splicing of CRS2-dependent introns (Figure 5). In addition, the CRS2–CAF complexes are bound tightly to their cognate group II introns in vivo, as shown by the ability of αCAF antibodies to co-immunoprecipitate CRS2 and their genetically defined target intron RNAs from chloroplast extract (Figures 4 and 6). The results argue that CRS2–CAF complexes bind specific introns to form functional splicing RNPs, with the CAF subunit determining the intron specificity of the complex.
CRM domains are a shared element in group II intron splicing factors
A striking feature of the CAFs is their sequence similarity with CRS1, a previously identified group II intron splicing factor in maize (Till et al., 2001). This similarity is limited to repeated 10 kDa segments found twice in each CAF and three times in CRS1 (Figure 3). These same segments in CRS1 previously were noted to be related to a conserved ORF represented in eubacteria and archaea (Till et al., 2001). The prokaryotic members of this family have been designated as uncharacterized protein family (UPF) 0044 in the Pfam database. Recently, we found that the E.coli member of this family, YhbY, is bound specifically to pre-50S ribosomal subunits, suggesting a role in ribosome maturation (T.Kawamura and A.Barkan, in preparation). Here, we name this domain the chloroplast RNA splicing and ribosome maturation (CRM) domain to reflect the functions established for the four characterized members of the family (CRS1, CAF1, CAF2 and YhbY).
There is considerable evidence that CRM domains bind RNA. First, all four of the characterized CRM domain proteins are components of RNP complexes in vivo. Secondly, purified, recombinant CRS1 binds its cognate intron, the atpF intron, with high affinity and specificity in vitro (O.Ostersetzer and A.Barkan, in preparation). Finally, the crystal structures of E.coli YhbY and its predicted ortholog in Haemophilus influenzae revealed structural similarity to known RNA-binding proteins and a putative RNA-binding surface (Ostheimer et al., 2002; Willis et al., 2002). The CRM domains in CAF1, CAF2 and CRS1 are expected to adopt a structure similar to that of YhbY and to share a similar RNA-binding surface (Ostheimer et al., 2002).
These findings clarify the biochemical basis for the intron specificity of CRS2. Nine of the 17 group II introns in the chloroplast require CRS2 for splicing (Jenkins et al., 1997), and CRS2 is found in chloroplast RNP particles that co-sediment with several of its intron targets (Jenkins and Barkan, 2001). However, recombinant CRS2 binds poorly and without specificity to its cognate group II intron RNAs in vitro (data not shown). As such, it has been unclear how CRS2 recognizes its target introns in vivo. The results presented here suggest that the CAF subunit of each CRS2–CAF complex recruits CRS2 to the relevant introns and is responsible for the intron specificity of the complex. The phenotypes of the caf mutants show that the nine CRS2-dependent introns can be subdivided according to their requirement for either CAF1 or CAF2 (Figure 7). The co-immunoprecipitation data (Figure 6) show that these genetically defined intron specificities reflect preferential binding of each CAF to distinct intron subsets in vivo. CAF1 and CAF2 each harbor two CRM domains and are therefore predicted to interact directly with RNA. Indeed, purified recombinant CRS1, a protein with three CRM domains, binds atpF intron RNA with high affinity and specificity in vitro (O.Ostersetzer and A.Barkan, in preparation), so it seems likely that an analogous situation will hold for CAF1 and CAF2.
An unanswered question concerns the basis for the specificity of CAF1 and CAF2 for different sets of subgroup IIB introns. The putative RNA-binding regions of CAF1 and CAF2 (their CRM domains and flanking regions) are highly similar (see Figure 2). Furthermore, there are no obvious sequence motifs that distinguish the CAF1-dependent introns from the CAF2-dependent introns. In fact, the co-immunoprecipitation data suggest that, although CAF1 and CAF2 bind preferentially to different intron subsets, there is, nonetheless, some overlap in their substrate set in vivo (Figure 6). The features that distinguish CAF2 RNA substrates from CAF1 substrates may prove to be subtle.
CRS2–CAF complexes and intron folding
Both group I and group II introns consist of discrete elements of secondary structure that assemble into a compact, catalytically active conformation through the formation of inter- and intra-domain tertiary interactions (Qin and Pyle, 1998). The folding of these and other highly structured RNAs is complicated by their propensity to form incorrect but stable base pairings, which act as kinetic traps (Herschlag, 1995). In addition, the weakness of their tertiary interactions can make it difficult for the RNA to adopt and/or maintain the catalytically competent fold, as has been observed for the folding of a model group II intron in vitro (Swisher et al., 2002).
Proteins that facilitate the splicing of group I and group II introns could, in principle, compensate for both aspects of the RNA folding problem. Proteins that bind weakly and non-specifically to single-stranded RNA have been proposed to chaperone RNA folding by favoring the unfolded state to facilitate escape from kinetic traps (Herschlag, 1995). Natural intron substrates of proteins with ‘RNA chaperone’ activity have not been identified, but an enhancement of group I intron activity by RNA chaperones has been observed in artificial systems (Coetzee et al., 1994; Clodi et al., 1999; Waldsich et al., 2002). DEAD-box helicases facilitate the splicing of both group I (Mohr et al., 2002) and group II introns (Seraphin et al., 1989), presumably through the ATP-dependent disruption of mispaired regions.
Proteins that bind tightly to specific intron segments could promote the correct intron folding pathway by stabilizing a folding intermediate, inhibiting an off-pathway fold or stabilizing the active intron structure (Weeks, 1997). A number of group I and group II splicing factors have been characterized that bind tightly and specifically to their intron targets. Examples include group I intron maturases (Ho and Waring, 1999; Bassi et al., 2002; Solem et al., 2002), the group I intron host-encoded factors CBP2 (Weeks and Cech, 1995), CYT-18 (Guo and Lambowitz, 1992) and Mrs1 (Bassi et al., 2002), the group II intron-encoded maturase LtrA (Matsuura et al., 1997), and the group II intron host-encoded factors CRS1 (Till et al., 2001; O.Ostersetzer and A.Barkan, in preparation) and Raa3 (Rivier et al., 2001). The fact that the αCAF antisera efficiently co-immunoprecipitated both CRS2 and their cognate introns from chloroplast extract demonstrates the existence of stable and specific CRS2–CAF–intron complexes. Therefore, the CRS2–CAF complexes are likely to facilitate splicing at least in part by recognizing correctly folded elements of their intron targets. It will be interesting to determine the stage of the intron folding process at which the CRS2–CAF complex binds intron RNA and whether additional host-encoded proteins contribute to the splicing of CRS2–CAF-dependent introns.
CRM domains as ancient RNA-binding modules
Many host-encoded proteins that facilitate group I or group II intron splicing appear to have evolved from proteins that play fundamental roles in nucleic acid metabolism. A pseudouridine synthase and a PTH evolved into the chloroplast group II intron splicing factors Maa2 (Perron et al., 1999) and CRS2 (Jenkins et al., 2001), respectively. The mitochondrial group I intron splicing factors NAM2 and CYT-18 evolved from tRNA synthetases (reviewed in Lambowitz et al., 1999), and the group I splicing factor MRS1 is descended from a DNA junction-resolving enzyme (Wardleworth et al., 2000; Bassi et al., 2002). CRS1 and the CAFs provide additional examples of this general phenomenon. The CRM domains in these proteins are related to a conserved prokaryotic protein, whose E.coli representative, YhbY, is a pre-50S ribosome-binding protein (T.Kawamura and A.Barkan, in preparation). Thus, it appears that a single-domain protein that evolved in the biological context of ribosome assembly was recruited and expanded during the evolution of plant genomes to facilitate the assembly of other catalytic RNPs, namely group II intron RNPs in chloroplasts and, perhaps, mitochondria (see below). It will be interesting to determine whether there are structural similarities between the RNA motif recognized by YhbY and those recognized by the CRM domains of CRS1, CAF1 and CAF2.
The CRM domain family in plants
CAF1, CAF2 and CRS1 are members of a family of plant proteins defined by the presence of one or more CRM domains (Till et al., 2001). We have detected 16 predicted proteins with CRM domains in the A.thaliana genome (http://mips.gsf.de/proj/thal/db/index.html) and a similar set in the rice genome (http://www.tmri.org). Other than CRS1, CAF1 and CAF2, these proteins are uncharacterized, but it seems likely that they all interact with RNA. Plant CRM domains are found in diverse sequence contexts and are predicted to reside not only in the chloroplast, but also in the mitochondrion and the nucleo/cytoplasmic compartment. The diversity of the CRM domain family in plants suggests a diverse set of RNA targets. We have not detected sequences encoding CRM domains in animal or fungal genomes.
Four Arabidopsis proteins are particularly similar to maize CAF1 and CAF2. We tentatively assign two of these proteins, At2g20020 and At1g23400, as the orthologs of CAF1 and CAF2, respectively, because of their predicted chloroplast localization, their extensive overall sequence identities and their sharing of characteristic sequences at their C-termini (Figure 2). The other two CAF-like proteins in Arabidopsis, At4g31010 and At5g54890, are predicted to be targeted to the mitochondrion. The Arabidopsis genome also encodes a predicted mitochondrially localized CRS2-like protein. This raises the intriguing possibility that the splicing of the group II introns in plant mitochondria involves proteins that are related to CRS2 and the CAFs. In Nicotiana sylvestris, the nuclear gene NMS1 is required for the splicing of the group II intron in the mitochondrial nad4 pre-mRNA (Brangeon et al., 2000). The cloning of NMS1 has not been reported, but it will be interesting to learn whether it encodes a CRS2 or CAF homolog.
The C.reinhardtii chloroplast genome harbors two group II introns, both transcribed in pieces and trans-spliced. Mutations in at least 14 nuclear genes disrupt the splicing of one or both of those introns (Goldschmidt-Clermont et al., 1990), and the molecular cloning of two such genes has been reported: Maa2 has homology to pseudouridine synthases (Perron et al., 1999) and Raa3 is a novel protein that is found in an RNP complex containing its target intron (Rivier et al., 2001). These proteins are unrelated to CRS1, CRS2, CAF1 or CAF2 (the only known host factors for group II intron splicing in land plants). However, we have detected a C.reinhardtii expressed sequence tag encoding a CRM domain (unpublished observations), leaving open the possibility that there may be similarities between the plastid splicing machineries in plants and green algae. The requirement of chloroplast group II introns for nucleus-encoded protein cofactors raises the intriguing possibility that the regulated synthesis or activity of these proteins could regulate the splicing of chloroplast introns. In fact, the tissue-dependent splicing of several group II introns in the maize chloroplast has been reported (Barkan, 1989). As such, the splicing factors described here could mediate the regulation of chloroplast group II intron splicing and contribute to the regulation of chloroplast biogenesis and function.
Materials and methods
Yeast two-hybrid screen
The yeast two-hybrid screen was performed with the Stratagene HybriZAP-2.1 Two-Hybrid Vector system following the manufacturer’s instructions. The CRS2 ORF, lacking sequences encoding the predicted chloroplast targeting sequence, was amplified by PCR using the primers CRS2D (GCGGAATTCATGGAATACACGCCC) and CRS2L (GGA GGTCGACTTCAAACCCTG). The product was cloned into the EcoRI and SalI sites of the pBD-Gal4 Cam plasmid, to generate the plasmid pBD-CRS2. The maize cDNA library was generated from leaf RNA extracted from 14-day-old seedlings (inbred line B73, Pioneer HiBred) grown in light–dark cycles. cDNAs were inserted into Stratagene’s Hybrizap-2.1 vector according to the manufacturer’s instructions.
Antibodies to CAF1 and CAF2
A CAF2-specific peptide antigen corresponding to residues 564–583 (GNEEGQLEQSPDLRDDEHFD) was synthesized and used for immunizing rabbits at Alpha Diagnostic International. Recombinant CAF1-specific antigen (residues 398–460) was generated by expression in E.coli. Crude sera were affinity purified against the same antigen. Each affinity-purified antibody preparation detected one predominant protein on immunoblots of chloroplast stroma, and these proteins corresponded in size to that predicted for the cognate antigen.
Co-immunoprecipitation of CRS2–CAF–intron complexes from chloroplast extract
Intact chloroplasts were isolated from the leaves of maize seedlings as described previously (Jenkins and Barkan, 2001). Chloroplasts were lysed by incubation on ice in a minimal volume of hypotonic lysis buffer [30 mM HEPES–KOH pH 7.7, 10 mM Mg acetate, 60 mM K acetate, 2 mM dithiothreitol (DTT) and a cocktail of protease inhibitors]. After a 30 min incubation punctuated by several rounds of vortexing, membranes were pelleted by centrifugation for 30 min at 29 000 r.p.m. in a Beckman OptimaTL ultracentrifuge (TLA 100.2 rotor, 36 500 g). The supernatant constitutes the stromal fraction used for these experiments.
Formalin-fixed Staphylococcus aureus cells (IgSorb; The Enzyme Co.) were washed three times in co-IP buffer (150 mM NaCl, 20 mM Tris–HCl pH 7.5, 1 mM EDTA, 0.5% NP-40, 5 µg/ml aprotinin) and resuspended in co-IP buffer to a concentration of 10% (v/v). A 1 mg aliquot of stromal protein (∼100 µl of extract) was pre-cleared by incubation with 100 µl of washed S.aureus cell suspension for 5 min on ice, followed by brief microcentrifugation to pellet the cells. Affinity-purified antisera were added to the pre-cleared stroma and incubated on ice for 1 h. A 100 µl aliquot of washed S.aureus cell suspension was then added to each reaction and incubated on ice for 1 h with rocking. The cells were pelleted by microcentrifugation for 1 min; the resulting supernatants constitute the supernatant fraction in Figure 6. Pellets were washed three times by resuspension in 0.5 ml of co-IP buffer and microcentrifugation for 1 min. The final washed pellet was suspended in a small volume of co-IP buffer: one-quarter of this suspension was used for western analysis of immunoprecipitated proteins (Figure 4), and the remainder was used for RNA extraction. RNA was extracted from the supernatant and pellet fractions using phenol–chloroform, after the addition of SDS to 1%, EDTA to 5 mM and 2 µg of yeast tRNA. Equal proportions of each RNA sample were applied to a nylon membrane through a slot-blot manifold, and hybridized with radiolabeled DNA probes specific for the indicated intron sequences. Probes were generated by PCR amplification of the relevant chloroplast sequences, and radiolabeled by the random priming method.
Recovery of caf1 and caf2 mutants in a reverse genetic screen
The reverse genetic resource and screening method are described at http://chloroplast.uoregon.edu. In brief, pooled DNA samples from a collection of ∼2000 Mu transposon-induced non-photosynthetic maize mutants were screened by PCR, with a Mu primer in conjunction with a caf1- or caf2-specific primer. Gene primers had the following sequences: caf1, CGTTTGGATTTGAGGCTCCC; caf2, GTTGTGTGAAATGTGCGGCTTGA. Amplification of caf sequences occurs when an individual in a pool has a Mu transposon inserted near sequences bound by the gene-specific primer. Amplification of caf sequences was detected by Southern hybridization of PCR products, using caf1 or caf2 cDNA probes. Positive individuals within positive pools were then identified in an analogous fashion. Mu insertion sites were determined by DNA sequence analysis of the PCR products, and are shown in the context of the protein sequences in Figure 2. The sites of the Mu insertions in the caf1 mutants are 5′-GCCCGCTTGCCCT (Mu8 in caf1-1) and CTCGCTCCTTCCT (Mu3 in caf1-2) GTCCCCGGCT-3′. The site of the Mu insertion in the caf2-1 mutant is 5′-AGGGTGAGCCCC (Mu1) GGTGACGGCAGG-3′.
The mutations were propagated by crossing heterozygous siblings of each mutant to inbred lines and subsequent self-pollination to recover homozygous mutants. Genotypes were determined by PCR with Mu and gene-specific primers (as above) and by genomic Southern blot analysis.
RNase-protection and northern blot analyses
Total leaf RNA was extracted with TRIzol reagent (Bethesda Research Laboratories) according to the manufacturer’s instructions. crs2-1 and caf2-1 RNA was extracted from homozygous mutants. caf1 mutant tissue came from a cross between caf1-1/+ and caf1-2/+ plants, and is therefore caf1-1/caf1-2. iojap RNA came from fully-albino homozygous mutant seedling leaves. The RNase protection protocol, northern blot protocol and probes have been described previously (Jenkins et al., 1997). RNase protection assays used 6 µg of wild-type leaf RNA or 10 µg of mutant leaf RNA.
Acknowledgments
Acknowledgements
We thank Brian W.Matthews, Oren Ostersetzer and Kenny Watkins for useful discussions and comments on the manuscript. This work was supported by grants to A.B. from the National Science Foundation (DBI 0077756 and MCB-9904666) and from the NIH to B.W.Matthews (GM20066). G.J.O. was supported in part by NIH Training Grant GM07759.
References
- Barkan A. (1989) Tissue-dependent plastid RNA splicing in maize. Plant Cell, 1, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. and Martienssen,R.A. (1991) Inactivation of maize transposon Mu suppresses a mutant phenotype by activating an outward-reading promoter near the end of Mu1. Proc. Natl Acad. Sci. USA, 88, 3502–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi G.S., de Oliviera,D.M., White,M.F. and Weeks,K.M. (2002) Recruitment of intron-encoded and co-opted proteins in splicing of the bI3 group I intron RNA. Proc. Natl Acad. Sci. USA, 99, 128–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen L. and Vogel,J. (2001) The ins and outs of group II introns. Trends Genet., 17, 322–331. [DOI] [PubMed] [Google Scholar]
- Brangeon J. et al. (2000) Defective splicing of the first nad4 intron is associated with lack of several complex I subunits in the Nicotinia sylvestris NMS1 nuclear mutant. Plant J., 21, 269–280. [DOI] [PubMed] [Google Scholar]
- Cech T.R. (1986) The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell, 44, 207–210. [DOI] [PubMed] [Google Scholar]
- Clodi E., Semrad,K. and Schroeder,R. (1999) Assaying RNA chaperone activity in vivo using a novel RNA folding trap. EMBO J., 18, 3776–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee T., Herschlag,D. and Belfort,M. (1994) Eschericia coli proteins, including ribosomal protein S12, facilitate in vitro splicing of phage T4 introns by acting as RNA chaperones. Genes Dev., 8, 1575–1588. [DOI] [PubMed] [Google Scholar]
- Collins C.A. and Guthrie,C. (2000) The question remains: is the spliceosome a ribozyme. Nat. Struct. Biol., 7, 850–854. [DOI] [PubMed] [Google Scholar]
- Dai L. and Zimmerly,S. (2002) Compilation and analysis of group II intron insertions in bacterial genomes: evidence for retroelement behavior. Nucleic Acids Res., 30, 1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L. and Zimmerly,S. (2003) ORF-less and reverse-transcriptase-encoding group II introns in achaebacteria, with a pattern of homing into related group II intron ORFs. RNA, 9, 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush T.H. (1999) Mobile introns: retrohoming by complete reverse splicing. Curr. Biol., 9, R11–R14. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen,H., Brunak,S. and von Heijne,G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol., 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Fields S. and Song,O. (1989) A novel genetic system to detect protein–protein interactions. Nature, 340, 245–246. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M., Girard-Bascou,J., Choquet,Y. and Rochaix,J.-D. (1990) Trans-splicing mutants of Chlamydomonas reinhardtii. Mol. Gen. Genet., 223, 417–425. [DOI] [PubMed] [Google Scholar]
- Gordon P.M. and Piccirilli,J. (2001) Metal ion coordination by the AGC triad in domain 5 contributes to group II intron catalysis. Nat. Struct. Biol., 8, 893–898. [DOI] [PubMed] [Google Scholar]
- Gordon P.M., Sontheimer,E.J. and Piccirilli,J.A. (2000) Metal ion catalysis during the exon-ligation step of nuclear pre-mRNA splicing: extending the parallels between the spliceosome and group II introns. RNA, 6, 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q. and Lambowitz,A.M. (1992) A tyrosyl-tRNA synthetase binds specifically to the group I intron catalytic core. Genes Dev., 6, 1357–1372. [DOI] [PubMed] [Google Scholar]
- Herschlag D. (1995) RNA chaperones and the RNA folding problem. J. Biol. Chem., 270, 20871–20874. [DOI] [PubMed] [Google Scholar]
- Higgins D., Thompson,J., Gibson,T., Thompson,J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y. and Waring,R. (1999) The maturase encoded by a group I intron from Aspergillus nidulans stabilizes RNA tertiary structure and promotes rapid splicing. J. Mol. Biol., 292, 987–1001. [DOI] [PubMed] [Google Scholar]
- Jenkins B.D. and Barkan,A. (2001) Recruitment of a peptidyl-tRNA hydrolase as a facilitator of group II intron splicing in chloroplasts. EMBO J., 20, 872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins B.D., Kulhanek,D.J. and Barkan,A. (1997) Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell, 9, 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz A.M., Caprara,M.G., Zimmerly,S. and Perlman,P.S. (1999) Group I and group II ribozymes as RNPs: clues to the past and guides to the future. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 451–485. [Google Scholar]
- Matsuura M. et al. (1997) A bacterial group II intron encoding reverse transcriptase, maturase and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev., 11, 2910–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menninger J. (1976) Peptidyl-transfer RNA dissociates during protein synthesis from ribosomes of Eschericia coli. J. Biol. Chem., 251, 3392–3398. [PubMed] [Google Scholar]
- Michel F. and Ferat,J.-L. (1995) Structure and actvities of group II introns. Annu. Rev. Biochem., 64, 435–461. [DOI] [PubMed] [Google Scholar]
- Michel F., Umesono,K. and Ozeki,H. (1989) Comparative and functional anatomy of group II catalytic introns—a review. Gene, 82, 5–30. [DOI] [PubMed] [Google Scholar]
- Mohr S., Stryker,J.M. and Lambowitz,A.M. (2002) A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell, 109, 769–779. [DOI] [PubMed] [Google Scholar]
- Nilsen T.W. (1998) RNA-RNA interactions in nuclear pre-mRNA splicing. In Simons,R.W. and Grunberg-Manago,M. (eds), RNA Structure and Function. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 279–307. [Google Scholar]
- Ostheimer G.J., Barkan,A. and Matthews,B.W. (2002) Crystal structure of E.coli YhbY: a representative of a novel class of RNA binding proteins. Structure, 10, 1593–1601. [DOI] [PubMed] [Google Scholar]
- Perron K., Goldschmidt-Clermont,M. and Rochaix,J.-D. (1999) A factor related to pseudouridine synthases is required for chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J., 18, 6481–6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P.Z. and Pyle,A.M. (1998) The architectural organization and mechanistic functions of group II intron structural elements. Curr. Opin. Struct. Biol., 8, 301–308. [DOI] [PubMed] [Google Scholar]
- Rivier C., Goldschmidt-Clermont,M. and Rochaix,J.-D. (2001) Identification of an RNA–protein complex involved in chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J., 20, 1765–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt E., Mechulam,Y., Fromant,M., Plateau,P. and Blanquet,S. (1997) Crystal structure at 1.2 Å resolution and active site mapping of Escherichia coli peptidyl-tRNA hydrolase. EMBO J., 16, 4760–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin B., Simon,M., Boulet,A. and Faye,G. (1989) Mitochondrial splicing requires a protein from a novel helicase family. Nature, 337, 84–87. [DOI] [PubMed] [Google Scholar]
- Sharp P.A. (1991) ‘Five easy pieces’. Science, 254, 663. [DOI] [PubMed] [Google Scholar]
- Solem A., Chatterjee,P. and Caprara,M. (2002) A novel mechanism for protein-assisted group I intron splicing. RNA, 8, 412–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer E.J., Sun,S. and Piccirilli,J. (1997) Metal ion catalysis during splicing of premessenger RNA. Nature, 388, 801–805. [DOI] [PubMed] [Google Scholar]
- Swisher J.F., Su,L.J., Brenowitz,M., Anderson,V.E. and Pyle,A.M. (2002) Productive folding to the native state by a group II intron ribozyme. J. Mol. Biol., 315, 297–310. [DOI] [PubMed] [Google Scholar]
- Till B., Schmitz-Linneweber,C., Williams-Carrier,R. and Barkan,A. (2001) CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA, 7, 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N., Hausner,G. and Zimmerly,S. (2001) Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA, 7, 1142–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro N. (2003) Bacteria and Archaea group II introns: additional mobile genetic elements in the envronment. Environ. Microbiol., 5, 143–151. [DOI] [PubMed] [Google Scholar]
- Valadkhan S. and Manley,J.L. (2001) Splicing-related catalysis by protein-free snRNAs. Nature, 413, 701–707. [DOI] [PubMed] [Google Scholar]
- Villa T., Pleiss,J.A. and Guthrie,C. (2002) Spliceosomal snRNAs: Mg2+-dependent chemistry at the catalytic core? Cell, 109, 149–152. [DOI] [PubMed] [Google Scholar]
- Vogel J., Borner,T. and Hess,W. (1999) Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plants mutants. Nucleic Acids Res., 27, 3866–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldsich C., Grossberger,R. and Schroeder,R. (2002) RNA chaperone StpA loosens interaction of the tertiary structure in the td group I intron in vivo. Genes Dev., 16, 2300–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardleworth B.J., Kvaratskhelia,M. and White,M.F. (2000) Site-directed mutagenesis of the yeast resolving enzyme CceI reveals catalytic residues and relationship with the intron-splicing factor Mrs1. J. Biol. Chem., 275, 23725–23728. [DOI] [PubMed] [Google Scholar]
- Weeks K.M. (1997) Protein-facilitated RNA folding. Curr. Opin. Struct. Biol., 7, 336–342. [DOI] [PubMed] [Google Scholar]
- Weeks K.M. and Cech,T.R. (1995) Efficient protein-facilitated splicing of the yeast mitochondrial bI5 intron. Biochemistry, 34, 7728–7738. [DOI] [PubMed] [Google Scholar]
- Willis M.A., Krajewski,W., Chalamasetty,V.R., Retty,P.,A.,H. and Herzberg,O. (2002) Structure of HI1333 (YhbY), a putative RNA-binding protein from Haemophilus influenzae. Proteins, 49, 423–426. [DOI] [PubMed] [Google Scholar]