Abstract
E2F transcription factors can activate or actively repress transcription of their target genes. The role of active repression during normal development has not been analyzed in detail. dE2F1su89 is a novel allele of dE2F1 that disrupts dE2F1’s association with RBF [the Drosophila retinoblastoma protein (Rb) homolog] but retains its transcription activation function. Interestingly, the dE2F1su89 mutant, which has E2F activation by dE2F1su89 and active repression by dE2F2, is viable and fertile with no gross developmental defects. In contrast, complete removal of active repression in de2f2;dE2F1su89 mutants results in severe developmental defects in tissues with extensive endocycles but not in tissues derived from mitotic cycles. We show that the endoreplication defect resulted from a failure to downregulate the level of cyclin E during the gap phase of the endocycling cells. Importantly, reducing the gene dosage of cyclin E partially suppressed all the phenotypes associated with the endoreplication defect. These observations point to an important role for E2F–Rb complexes in the downregulation of cyclin E during the gap phase of endocycling cells in Drosophila development.
Keywords: active repression/E2F/endoreplication/genetic screen/Rb
Introduction
The E2F transcription factors, which consist of a subunit of the E2F family of proteins and a subunit of the DP proteins, are important regulators of S phase and are key targets of the retinoblastoma (Rb) family of proteins. In addition to activating transcription and promoting cell cycle progression, E2F transcription factors can also negatively regulate cell cycle progression by actively repressing target gene expression through their association with Rb family proteins (Weintraub et al., 1992, 1995; Zhang et al., 1999; Gaubatz et al., 2000; He et al., 2000). Six E2F and two DP proteins have been identified in mammalian systems (Dyson, 1998; Trimarchi and Lees, 2002). Extensive studies suggest that these six mammalian E2F proteins can be divided into three classes: the activating E2Fs (E2F-1, E2F-2 and E2F-3), the repressive E2Fs (E2F-4 and E2F-5) and E2F-6, which was shown to be a component of the mammalian Bmi1-containing polycomb complex (Trimarchi et al., 2001). The activating E2Fs function mainly to activate transcription, although this class of E2F proteins may also contribute to the repression of E2F target genes. In contrast, the repressive E2Fs function primarily to mediate repression. Although a role for E2F–Rb complexes in active repression and in maintaining G1 arrest is well established in cell culture studies (Weintraub et al., 1992, 1995; Zhang et al., 1999), it is not clear what the biological consequences will be when all E2F–Rb complexes are removed during normal development. Such questions are difficult to address in mammalian systems due to the presence of extended family members of E2F, Rb and DP proteins.
The E2F, DP and Rb families of proteins are conserved between Drosophila and mammalian systems (for reviews see Dyson, 1998; Trimarchi and Lees, 2002). In contrast to the extended family members of the E2F and DP gene families, only two E2F genes and one DP gene have been identified in the Drosophila genome (Dynlacht et al., 1994; Ohtani and Nevins, 1994; Sawado et al., 1998). Studies of the roles of dE2F1, dE2F2 and dDP have revealed that dE2F1 and dDP proteins are essential for the E2F-dependent G1–S transcription program and for cell proliferation in several developmental contexts (Duronio and O’Farrell, 1995; Duronio et al., 1995, 1996; Brook et al., 1996; Royzman et al., 1997; Du, 2000). In contrast, dE2F2 is not absolutely required during Drosophila development but functions mainly as a transcription repressor by recruiting Rb family proteins to the E2F target genes (Frolov et al., 2001; Stevaux et al., 2002), and it plays a role in regulating the transition from genomic replication to amplification in late stage follicle cells (Cayirlioglu et al., 2001). These observations indicate that the two Drosophila E2F proteins appear to behave like the two different classes of E2Fs of their mammalian counterparts: dE2F1 functions mainly as a transcriptional activator (Du, 2000), similar to the activating E2Fs (E2F1–3), while dE2F2 functions mainly as a corepressor of RBF, similar to the repressive E2F (E2F4 and 5) in mammalian systems (Frolov et al., 2001; Stevaux et al., 2002). The simplified and yet conserved function and regulation of the E2F–Rb pathway makes Drosophila an ideal system to characterize the roles of the E2F–Rb complexes during normal development.
In this report, we describe a novel gain-of-function allele of dE2F1, dE2F1su89, which uncouples the ability of dE2F1 to activate transcription from its ability to interact with RBF. The dE2F1su89 mutants, in which E2F activation and active repression were mediated by dE2F1su89 and dE2F2, respectively, were viable and fertile, with no gross developmental defect. Removal of dE2F2 in the dE2F1su89 mutant background led to complete removal of active repression by E2F proteins. The majority of de2f2;dE2F1su89 mutants died during development and showed extensive defects in endocycle tissues. We show that the endoreplication defects are caused by a failure to downregulate cyclin E during the gap phase of the endocycling cells in the absence of active repression by E2F–Rb complexes. Importantly, reducing the gene dosage of cyclin E partially suppressed all the phenotypes associated with the endoreplication defect. These observations point to an important role for E2F–Rb complexes in the downregulation of cyclin E during the gap phase of endocycling cells in normal development.
Results
A special gain-of-function mutation in the dE2F1 gene
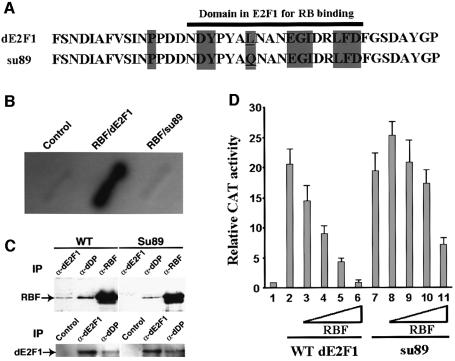
We identified a novel allele of dE2F1, dE2F1su89, from a genetic screen for suppressors of the RBF overexpression phenotype (see Supplementary data available at The EMBO Journal Online). Sequence analysis revealed that dE2F1su89 contains a single base pair mutation in the conserved Rb-binding domain that converts the conserved amino acid leucine at position 786 to glutamine (Figure 1A). To test whether this mutation disrupts the interaction between RBF and dE2F1, a yeast two-hybrid interaction assay was performed. As shown in Figure 1B, dE2F1su89 can no longer bind to RBF. To demonstrate further the effect of this mutation with endogenous proteins, a co-immunoprecipitation experiment was carried out. While both dE2F1 and dDP were co-immunoprecipitated with RBF from wild-type embryo extracts, no dE2F1 was co-immunoprecipitated with RBF from the dE2F1su89 embryo extracts, even though similar levels of dE2F1 protein were present in the two extracts (Figure 1C). These results indicate that the endogenous dE2F1su89 and RBF proteins did not form a complex. Interestingly, dDP was still co-immunoprecipitated with RBF from the dE2F1su89 embryo extracts, indicating that dDP can still form a complex with RBF in the dE2F1su89 mutant background, probably through the other Drosophila E2F protein, dE2F2.
Fig. 1. Molecular characterization of the dE2F1su89 mutation. (A) The point mutation (underlined) in the dE2F1su89 C-terminus is shown. Amino acids that are conserved between Drosophila and mammalian E2Fs are shaded in gray, and the Rb-binding domain of dE2F1 is indicated with a black bar. (B) dE2F1su89 did not interact with RBF in a yeast two-hybrid interaction assay. β-Gal activity on patches of yeast transformed with control plasmids or plasmids encoding RBF, wild-type dE2F1 or dE2F1su89 (su89) is shown as indicated. (C) Endogenous dE2F1su89 protein did not form a complex with RBF. Extracts from wild-type (WT) or dE2F1su89 (su89) embryos were immunoprecipitated with anti-dE2F1, dDP and RBF antibodies as indicated, followed by western blot with an antibody against RBF or dE2F1 as indicated to determine the association between the endogenous RBF and dE2F1su89 proteins. (D) dE2F1su89 can activate transcription normally but is defective in its regulation by RBF. SL2 cells were transfected with no dE2F1 (1), a constant amount of wild-type dE2F1 (2–6) or dE2F1su89 (7–11), and increasing amounts of RBF as indicated. The averages of normalized CAT activities from two independent experiments are shown.
As dE2F1su89 contains a point mutation in the RBF-binding domain that overlaps with the transcription activation domain, the ability of dE2F1su89 to activate transcription and its regulation by RBF was determined. As shown in Figure 1D, transfection of dE2F1su89 into Drosophila SL2 cells led to transcriptional activation from an E2F reporter construct to the same extent as transfection of wild-type dE2F1, indicating that the point mutation in dE2F1su89 did not affect its ability to activate transcription (Figure 1D, columns 2 and 7). However, this point mutation impaired the regulation of dE2F1su89 by RBF. Co-transfection of RBF effectively inhibited the transcriptional activation induced by wild-type dE2F1 in a dosage-dependent manner (Figure 1D, columns 2–6), and no transcriptional activation was observed at high levels of RBF (Figure 1D, column 6). In contrast, no significant inhibition of dE2F1su89-induced transcriptional activation was observed at low levels of RBF (Figure 1D, columns 7–10). At the highest level of RBF transfected, there was still ∼7- to 8-fold transcriptional activation observed (Figure 1D, column 11). These data suggest that dE2F1su89 is an allele of dE2F1 that can activate transcription normally but is defective in its regulation by RBF.
Genetic interactions between RBF and dE2F1su89
To test the effect of disrupting the interaction between dE2F1 and RBF on the consequences of RBF overexpression in vivo, we examined the effects of dE2F1su89 mutation on phenotypes induced by RBF overexpression, including E2F target gene expression, cell cycle regulation and the consequent developmental defects.
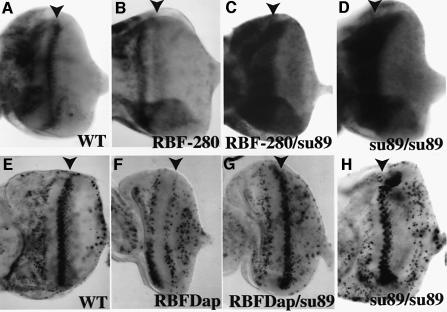
RBF-280, a form of RBF that has four consensus cdk sites mutated, cannot be regulated by cyclin E or cyclin D (Xin et al., 2002). Overexpression of wild-type RBF or RBF-280 in the posterior portion of the eye disc (GMRRBF4C and GMRRBF-2802) inhibited the expression of proliferating cell nuclear antigen (PCNA), an E2F target gene, in the developing eye (Figure 2B and data not shown). Introducing one copy of dE2F1su89 restored PCNA expression even in the presence of the constitutively active RBF-280 (Figure 2C). In addition, although the dE2F1su89 mutation does not significantly affect PCNA expression in the anterior region of the eye discs, a high level of PCNA expression was observed in the second mitotic wave as well as in the morphogenetic furrow of the dE2F1su89 discs and the GMRRBF-2802/ dE2F1su89 discs (Figure 2C and D). These results are consistent with the idea that RBF cannot inhibit dE2F1su89 in vivo.
Fig. 2. dE2F1su89 mutation blocked RBF-mediated inhibition of E2F target gene expression and S phase entry in the second mitotic wave (SMW). (A–D) In situ hybridization of PCNA, an E2F target gene, in eye discs of various genotypes is shown. The genotypes are (A) wild-type, (B) +/GMRRBF-2802, (C) dE2F1su89/GMRRBF-2802 and (D) dE2F1su89. Expression of RBF-280 in the dE2F1su89/+ background did not inhibit PCNA expression in SMW. Note that a high level of PCNA expression was observed in the furrow of the dE2F1su89/GMRRBF-2802 discs, an expression pattern that was similar to that of the dE2F1su89 discs (D). (E–H) BrdU incorporation by eye discs from various genotypes is shown. The genotypes are (E) wild-type, (F) +/GMRRBFDap, (G) dE2F1su89/GMRRBFDap and (H) dE2F1su89. GMRRBFDap expresses both RBF and Dap in the posterior part of the eye, which delayed and partially inhibited S phase in SMW (F). Introducing one copy of dE2F1su89 into a GMRRBFDap background suppressed the inhibition of S phase entry in SMW (G). Arrowheads indicate the second mitotic wave.
dE2F1su89 also suppressed the cell cycle defects associated with RBF overexpression. As reported previously, overexpression of either the wild-type RBF or the constitutively active RBF-280 alone did not inhibit S phase entry in the second mitotic wave of the developing eye (Xin et al., 2002), while expression of RBF together with the Drosophila p27 family cdk inhibitor Dacapo (GMRRBFDap) can inhibit or delay S phase entry in the second mitotic wave dependent upon the level of RBF and Dap expression (de Nooij et al., 1996; Xin et al., 2002; Figure 2F). As shown in Figure 2G, introducing one copy of the dE2F1su89 mutation restored normal S phase entry in the second mitotic wave even in the presence of RBF and Dacapo overexpression. Thus dE2F1su89 also suppressed the cell cycle effect of RBF overexpression in vivo.
Furthermore, dE2F1su89 suppressed all the developmental phenotypes caused by RBF overexpression. dE2F1su89 completely suppressed a relatively weak ommatidia fusion and missing bristle phenotype induced by overexpression of wild-type RBF alone (GMRRBF4C) as well as a much stronger phenotype induced by co-expression of RBF and Dacapo (GMRRBFDap) (Figure 3A–E). Interestingly, while a mutation in ptc, a negative regulator of the hh pathway that was shown to regulate cyclin D and cyclin E expression (Duman-Scheel et al., 2002), suppressed the GMRRBFDap phenotype (Figure 3F), it did not affect the phenotypes induced by RBF-280 expression (Figure 3G and I). These observations are consistent with the previous observation that RBF-280 cannot be regulated by cyclin D and cyclin E (Xin et al., 2002). Importantly, dE2F1su89 also suppressed the GMRRBF-2802 phenotype (Figure 3H). In addition, the ability of dE2F1su89 to suppress the effect of RBF overexpression is not limited to the eye. For example, overexpression of RBF in the wing disc using the dpp GAL4 driver inhibited a cross vein formation and reduced the inter-vein area (Figure 3K). These wing phenotypes were also completely suppressed by one copy of dE2F1su89 (Figure 3J–L).
Fig. 3. dE2F1su89-suppressed RBF overexpression induced developmental defects in the adult eyes and wings. SEM images of adult eyes from (A) wild-type, (B) GMRRBF4C, (D) GMRRBFDap and (G) GMRRBF-2802. Overexpression of RBF, RBF together with Dap, or RBF-280 led to different extents of the fused ommatidia and missing bristle phenotypes in the adult eyes. These phenotypes were suppressed by introducing one copy of dE2F1su89 into these backgrounds (C, E and H). Interestingly, while the ptc mutation suppressed the phenotypes of RBF and Dap overexpression (F), it did not suppress the phenotypes of GMRRBF-2802 (I). (J–L) dE2F1su89 also suppressed RBF overexpression-induced wing phenotypes. Expression of RBF in the wing using the dppGal4 driver blocked a cross vein formation and reduced the inter-vein area (K, indicated by arrows); these phenotypes were suppressed by the dE2F1su89 mutation (L).
Taken together, these results demonstrate that dE2F1su89 is a special allele of dE2F1 that retains its transcription activation function but disrupts its interaction with RBF. Importantly, dE2F1su89 suppressed all the phenotypes associated with RBF overexpression, indicating that dE2F1su89 can no longer be regulated by RBF in vivo.
dE2F1su89 flies are viable and fertile with no gross developmental defects
Recent studies suggest that dE2F1 functions mainly as a transcriptional activator (Du, 2000) while dE2F2 functions mainly as a transcriptional repressor by binding to RBF or RBF2 (Frolov et al., 2001; Stevaux et al., 2002). However, it is possible that the RBF–dE2F1 complex can also contribute to active repression. The identification of dE2F1su89, an allele of dE2F1 that separates the role of dE2F1 as a transcriptional activator from its potential role as a transcriptional repressor, provides a useful reagent to study the physiological consequence of unregulated dE2F1 activity in the presence or absence of dE2F2–RBF active repression complex during normal development.
Studies of rbf-null mutant flies have shown that RBF is required at multiple stages during Drosophila development. Most rbf-null mutant flies die during early larval development. The adult flies rescued with low-level expression of RBF show a number of alterations in adult structures, including rough eyes and abnormal macrochaetaes on the notum (Du, 2000). Since dE2F1 is a major downstream target of RBF that activates E2F target gene expression, we expected that dE2F1su89 flies would have phenotypes similar to those of rbf mutant flies. Surprisingly, dE2F1su89 homozygous flies were viable and fertile, with normal macrochaetaes on the notum (Figure 4A and B). dE2F1su89 mutant flies did have very slight eye phenotypes with extra bristles around some of the ommatidia (data not shown), similar to the weak eye phenotypes observed with flies carrying one copy of GMRdE2F1 and one copy of GMRdDP (Du et al., 1996b). Consistent with the reported endogenous pattern of dE2F1 in the eye (Brook et al., 1996) and the fact that dE2F1su89 protein did not bind RBF, high levels of PCNA expression were observed in the morphogenetic furrow and in areas immediately anterior as well as posterior to the furrow, including the second mitotic wave (Figure 2D). In addition, when compared with the wild-type eye discs, dE2F1su89 eye discs exhibit increased level of PCNA expression posterior to the second mitotic wave, although no obvious difference in PCNA expression was observed anterior to the furrow (Figure 2A and D). Bromodeoxy uridine (BrdU) staining revealed that dE2F1su89 eye discs have normal G1 arrest in the furrow (Figure 2H), indicating that the G1 arrest in the furrow is independent of E2F activity.
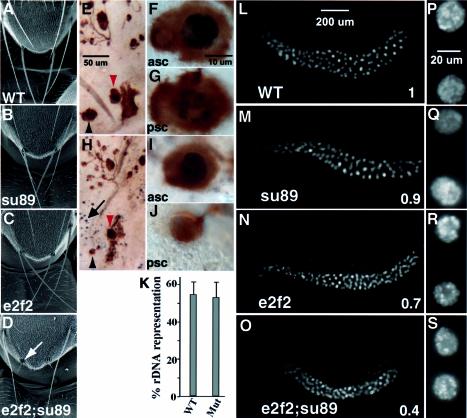
Fig. 4. Macrochaetae and salivary gland phenotypes of the de2f2;dE2F1su89 double mutants. SEM images of the fly notum are shown in (A–D). The wild-type (A), dE2F1su89 mutants (B) and de2f2 (C) showed normal macrochaetaes in the adults. In contrast, de2f2;dE2F1su89 adults showed severe defects in their macrochaetaes (D, white arrow). BrdU and mAb22C10 double labeling of the pupal nota is shown in (E–J). Pupa nota were dissected at 28 h AEL and were stained with mAb22C10 to mark the cell bodies of the sensory cluster (in brown) and BrdU antibody to visualize S phase cells (in black). A wild-type notum is shown in (E–G). (E) Low magnification view. The anterior scutellar bristle (asc, red arrowhead) and the posterior scutellar bristle (psc, black arrowhead) are shown in (F) and (G), respectively. The same developmental stage de2f2;de2f1su89 notum is show in (H–J). (H) Low magnification view. (I and J) High magnification views of the asc and psc, respectively. (K) The relative copy numbers of the rDNA sequences in wild-type and de2f2;dE2F1su89 mutant salivary glands are shown. The ratios of the hybridization signals of the rDNA sequences to a euchromatic sequencs from the salivary gland DNA were determined. Mitotically active diploid disc DNA, which is considered to have 100% representation of all the heterochromatic sequences, was used for normalization. The averages of at least three experiments are shown. WT, wild-type; Mut, de2f2;dE2F1su89 mutants. (L–S) Images of salivary glands stained with DAPI are shown. Low (L–O) and high (P–S) magnification images of salivary glands from wild-type (L and P), dE2F1su89 mutants (M and Q), de2f2 mutants (N and R) and de2f2;dE2F1su89 mutants (O and S). Numbers in (L–O) indicate the average DNA content of the salivary gland nuclei normalized by that from the wild-type. Note that while the salivary gland nuclei from the dE2F1su89 mutant had a similar DNA content to those from the wild-type (P = 0.24), the DNA content of the de2f2 mutant salivary gland nuclei was less than that of the wild-type (P < 0.00001) but more than that of the de2f2;dE2F1su89 (P < 0.00001).
One possible explanation for the lack of severe developmental defects in the dE2F1su89 mutant is the presence of active repression by dE2F2. To determine the role of E2F active repression during development, we removed dE2F2 in the dE2F1su89 mutant background to generate de2f2;dE2F1su89 mutants, in which the transcription activation function was retained while the active repression function of E2F was completely removed.
Removal of dE2F2 in the dE2F1su89 mutant background results in significant lethality and defective macrochaetaes
We generated a new de2f2 null allele, de2f2p111-5 (see Supplementary data). Trans-heterozygotes of de2f2p111-5 and the previously reported de2f276Q.1 (Frolov et al., 2001) were used in this study. Removing dE2F2 in the dE2F1su89 mutant background leads to lethality in the majority of de2f2;dE2F1su89 mutants. Of 145 de2f2;dE2F1su89 second or early third instar larvae that were followed, 75% of the mutants died during the larvae stage, 24% died during the pupae stage and only 1% of de2f2;dE2F1su89 larvae survived to adulthood. As expected from the lethality study, larval development of de2f2;dE2F1su89 mutants was very asynchronous: while most of the de2f2;dE2F1su89 larvae were smaller, some healthy de2f2;dE2F1su89 larvae can reach sizes similar to wild-type larvae, and their development was only delayed by about a day. The surviving de2f2;dE2F1su89 adults did not show gross defects in their adult structures except that the majority of them (88% from a total of 73) displayed extremely short macrochaetaes on the notum (Figure 4D). In contrast, both the dE2F1su89 and the de2f2 mutants had normal macrochaetaes in the adults (Figure 4A–C). The observed macrochaetae defects were similar to the phenotype observed in adult rbf mutant flies (Du, 2000). When examined under a scanning electronic microscope, these macrochaetaes were found to have formed the proper structure, but the shaft failed to grow to its normal size (Figure 4D, arrow).
de2f2;dE2F1su89 flies show defects in endoreplication during macrochaetae development
The adult sensilla of D.melanogaster are typically composed of one or more sensory neurons and three different accessory cells: the tormogen, trichogen and thecogen (Posakony, 1994). During pupal development, the trichogen cell and the tormorgen cell, which are responsible for producing the bristle shaft and the socket, respectively, become polypoid and increase in size through endoreplication. Since endoreplication contributes significantly to the cell size and the bristle size (Edgar and Orr-Weaver, 2001), we tested the possibility that the observed bristle defect in the de2f2;dE2F1su89 mutants is a consequence of defects in endoreplication.
Monoclonal antibody 22C10, which stains the cell body of all the cells in the sensory cluster (Hartenstein and Posakony, 1989), was used to identify the trichogen cells at the pupal stage. As shown in Figure 4E–J, the trichogen cells of de2f2;dE2F1su89 mutants were significantly smaller than those of the wild-type flies (Figure 4, compare brown staining of F, G and I, J). BrdU incorporation revealed that while the trichogen cells of the anterior scutellar bristle (asc) and posterior scutellar bristle (psc) were undergoing endoreplication in the scutella of the wild-type flies (Figure 4F and G, black staining), the psc trichogen nuclei of de2f2;dE2F1su89 mutant scutella was not detectably incorporating BrdU (Figure 4J). These results indicate that endoreplication in de2f2;dE2F1su89 macrochaetae cells is defective. Interestingly, while there was little BrdU staining in the non-sensory cluster cells in the wild-type scutella at this stage (Figure 4E), significant ectopic S phases were observed in the scutella of de2f2;dE2F1su89 mutant flies (Figure 4H, black staining indicated by an arrow). Thus de2f2;dE2F1su89 mutants also exhibited defects in maintaining cell cycle arrest in the scutella.
de2f2;dE2F1su89 double mutants show severe defects in salivary gland development
Endocycles are widely used during Drosophila development. Because cell size for a given cell type is generally proportional to the amount of nuclear DNA, endoreplication constitutes an effective strategy for growth of cells and tissues that are differentiated yet must continue to grow (Edgar and Orr-Weaver, 2001). The observed endoreplication defects in the de2f2;dE2F1su89 macrochaetaes prompted us carefully to examine another tissue with extensive endoreplication, the salivary gland, in de2f2;dE2F1su89 mutants.
The salivary glands from de2f2;dE2F1su89 third instar larvae were significantly smaller than those from dE2F1su89 or wild-type flies (Figure 4L–O). The number of nuclei in the de2f2;dE2F1su89 salivary gland (average = 127 nuclei/gland) was similar to that in wild-type salivary glands (average = 140 nuclei/gland, P = 0.07). Thus the smaller salivary gland in the de2f2;dE2F1su89 mutants was not due to a significantly decreased number of cells per salivary gland. In contrast, the distance between adjacent nuclei was greatly reduced in the double mutants (Figure 4P–S), indicating a significant reduction in the size of the cells in the de2f2;dE2F1su89 salivary glands.
The Drosophila salivary gland cells endoreplicate during embryonic and larval periods to produce giant polytene chromosomes and large cell size (Rudkin, 1972). A mature salivary gland cell contains up to 2048 copies of the euchromatic genome (Edgar and Orr-Weaver, 2001). To determine whether the small size of the de2f2; dE2F1su89 salivary gland cells was due to less DNA content, we quantified the salivary gland nuclei DNA from wandering third instar larvae with 4′,6-diamidino-2-phenylindole (DAPI) staining. While wild-type, dE2F1su89 and de2f2 reach late third instar at 120 h AEL (after egg lay), de2f2;dE2F1su89 mutants reach late third instar at 144 h AEL. As shown in Figure 4L–O, the salivary gland nuclei from late third instar de2f2;dE2F1su89 mutants had only 40% of the wild-type DNA content (P < 0.00001). In contrast, the DNA content of dE2F1su89 salivary gland nuclei was similar to that of wild-type (ratio = 0.9, P = 0.24). The DNA content of the de2f2 mutant salivary gland nuclei was less than that of the wild-type (ratio = 0.7, P < 0.00001), although it was still significantly more than that of the de2f2;dE2F1su89 double mutant (P < 0.00001).
The endocycles in Drosophila often have incomplete genomic replication during each round of S phase. As heterochromatic DNA is generally replicated late during each round of endoreplication, heterochromatin, which makes up ∼30% of the genome, is underreplicated in most polytene tissues (Lilly and Spradling, 1996). Thus there are two possible mechanisms that can lead to the decreased DNA content in the double mutant nuclei. One possibility is that there is less DNA replicated during each round of endoreplication in the double mutant cells than in the wild-type cells. Another possibility is that there are fewer rounds of DNA replication before pupation. As heterochromatic sequences generally replicate late, the first possibility would predict decreased heterochromatic sequence representation in the de2f2;dE2F1su89 mutant nuclei. To determine whether heterochromatic DNA representation was decreased in salivary gland DNA from de2f2;dE2F1su89 mutants compared with that from the wild-type, we examined the representation of a heterochromatic sequence, the rDNA sequence, in the wild-type and in de2f2;dE2F1su89 mutants as described previously (Lilly and Spradling, 1996). The ratio of the signal from the rDNA probe to the signal from a euchromatic probe was found to be the same between the de2f2;dE2F1su89 and wild-type salivary gland DNA (Figure 4K). Thus the decreased DNA content in the de2f2;dE2F1su89 mutants is not due to precocious shut off of DNA replication during each round of endoreplication but is probably due to fewer rounds of endocycles.
Since wild-type salivary glands start the endocycles 8.5 h AEL (Smith and Orr-Weaver, 1991) and complete endocycles at 120 h AEL with a DNA content of 2048 copies, the average length of each endocycle in wild-type salivary glands was ∼11 h. As de2f2;dE2F1su89 salivary gland cells had only 40% of the wild-type DNA content at 144 h AEL, the average length of each endocycle in de2f2;dE2F1su89 salivary glands was ∼16 h.
To determine whether the increased length of the endocycle is due to increased length of S phase or gap phase, we compared the proportion of cells that are in S phase and in gap phase between the wild-type and the de2f2;dE2F1su89 salivary glands. An average of 51 ± 10% nuclei were incorporating BrdU in the early third instar wild-type salivary glands. In contrast, only 26 ± 8% nuclei were incorporating BrdU from the same stage de2f2; dE2F1su89 salivary glands (P < 0.0003). Thus the estimated average lengths of S phase and gap phase of the wild-type salivary gland cells were 5.6 and 5.4 h, respectively. In contrast, the estimated average lengths of S phase and gap phase in the de2f2;dE2F1su89 salivary glands were 4.2 and 11.8 h, respectively. We conclude from these data that the increased length of endocycles in the de2f2;dE2F1su89 salivary glands is due to increased length of the gap phase.
Endoreplication defects in de2f2;dE2F1su89 salivary glands are caused by a failure to downregulate cyclin E in gap phase cells
Cyclin E is a key regulator of the endocycle. Oscillation of cyclin E-dependent kinase activity is required for multiple rounds of endoreplication (Edgar and Orr-Weaver, 2001). Cyclin E is required for entry into and progression through S phase of the endocycle (Knoblich et al., 1994). In addition, at the completion of one round of endoreplication, cyclin E activity needs to be downregulated, presumably for the re-loading of the replication origins for the next round of endoreplication. Continuous overexpression of cyclin E in Drosophila salivary glands inhibits endoreplication and results in a small salivary gland phenotype (Follette et al., 1998; Weiss et al., 1998).
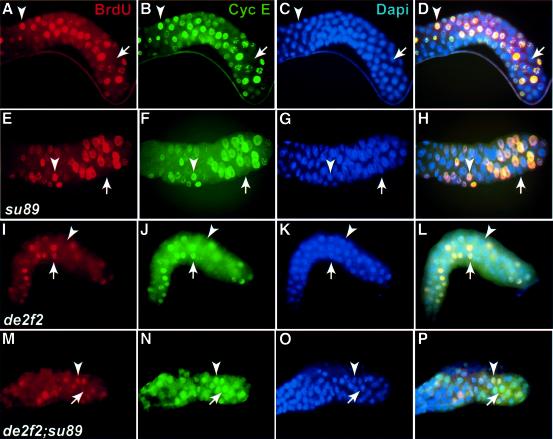
To determine whether cyclin E oscillation is defective in the de2f2;dE2F1su89 mutants, we monitored the correlation between DNA synthesis and cyclin E protein level by BrdU and cyclin E staining in wild-type, dE2F1su89, de2f2 and de2f2;dE2F1su89 double mutant salivary glands. As shown in Figure 5A–D, cyclin E expression (green) correlated perfectly with DNA synthesis (red) in a wild-type salivary gland. Only S phase cells exhibit a high level of cyclin E protein, and no cyclin E staining was observed in gap phase nuclei (Figure 5A–D, see nuclei indicated by arrows and arrowheads). Similar correlations between cyclin E level and BrdU incorporation were also observed in dE2F1su89 and de2f2 salivary glands (Figure 5E–L). In contrast, this correlation between BrdU and cyclin E staining was not observed in the de2f2;dE2F1su89 salivary glands (Figure 5M–P). While all the BrdU-positive cells had high cyclin E levels, high levels of cyclin E protein were also detected in a significant number of BrdU-negative cells (Figure 5M–P, see the nuclei indicated by the arrows). These results suggest that active repression by the E2F–Rb complexes is required for the downregulation of cyclin E during the gap phase of endocycles. In support of this idea, the level of cyclin E mRNA in de2f2;dE2F1su89 salivary glands was ∼4-fold as high as that in wild-type salivary glands as determined by semi-quantitative RT–PCR. As down-regulation of cyclin E is probably required for the re-loading of replication origins (Follette et al., 1998), accumulation of cyclin E in the post-replicative cells will probably inhibit entry into the next round of the endocycle, which may contribute to the reduced endoreplication phenotype of the de2f2;dE2F1su89 mutant flies.
Fig. 5. Salivary glands from de2f2;dE2F1su89 mutants show defects in cyclin E oscillation. Images of salivary glands from the wild-type (A–D), dE2F1su89 (E–H), de2f2 (I–L) and de2f2;dE2F1su89 (M–P) stained with BrdU (red, A, E, I and M), cyclin E (green, B, F, J and N) and DAPI (blue, C, G, K and O) are shown. The merged images are shown in (D), (H), (L) and (P). In de2f2;dE2F1su89 salivary glands, a significant number of nuclei that were not incorporating BrdU had high levels of cyclin E protein (see nuclei indicated by an arrow in M–P).
Reducing the gene dosage of cyclin E partially suppressed the lethality, macrochaetae and salivary gland phenotypes of de2f2;dE2F1su89 mutants
The results discussed above suggest that the endoreplication defects observed in the de2f2;dE2F1su89 mutants are likely to be due to a failure to downregulate cyclin E effectively during the gap phase in endocycling cells. If this is true, reducing the gene dosage of cyclin E may make it easier to downregulate the level of cyclin E during the gap phase and result in a suppression of the endoreplication-related phenotypes. Consistent with this prediction, reducing the dosage of cyclin E by half partially suppressed the de2f2;dE2F1su89 salivary gland endoreplication defect; the cycE/+,de2f2;dE2F1su89 salivary gland nuclei have about 70% of the wild-type DNA content (Figure 6A–C). In addition, reducing the dosage of cyclin E also partially suppressed the lethality of de2f2; dE2F1su89 mutants. From 127 cycE/+,de2f2;dE2F1su89 second or early third instar larvae that were followed, 52% survived to the pupae stage (compared with 25% survival of the de2f2;dE2F1su89 mutants, P < 0.0001) and 9% survived to adulthood (compared with 1% survival of the de2f2;dE2F1su89 mutants, P = 0.008). Finally, reducing the dosage of cyclin E by half also strongly suppresses the macrochaetae phenotype; 99% of the cycE/+,de2f2; dE2F1su89 flies show normal macrochaetaes (Figure 6E), compared with 12% of the de2f2;dE2F1su89 (P < 0.0001). These results strongly support the idea that phenotypes such as the macrochaetae defects, the small salivary glands and the lethality observed in the de2f2;dE2F1su89 mutants result, at least in part, from an endoreplication defect caused by a failure to downregulate cyclin E properly during the gap phase.
Fig. 6. Reducing the dosage of cyclin E by introducing one copy of the cycEAR95 mutation into the de2f2;dE2F1su89 mutant background partially suppressed the salivary gland endoreplication defect and strongly suppressed the macrochaetae defect of the de2f2;dE2F1su89 mutants. (A–C) Images of the salivary glands from wild-type (A), de2f2;dE2F1su89 (B) and cyclin E/+,de2f2;dE2F1su89 (C) stained with DAPI. The number in (A–C) indicates the average DNA content of the salivary gland nuclei. The cyclin E/+,de2f2;dE2F1su89 salivary glands were larger and had higher DNA content in their nuclei than the de2f2;dE2F1su89 salivary glands. (D and E) The SEM images of the fly notum from a de2f2;dE2F1su89 mutant (D) and a cyclin E/+,de2f2;dE2F1su89 mutant (E). Arrows in (D) show defective macrochaetaes. While 88% of the de2f2;dE2F1su89 flies had defective macrochaetaes, 99% of the cyclin E/+,de2f2;dE2F1su89 flies showed normal macrochaetaes.
Discussion
Using a novel allele of dE2F1, dE2F1su89, that uncouples the ability of dE2F1 to activate transcription from its ability to bind RBF, we determined the developmental consequence of an activating mutation of E2F in the presence or absence of E2F–Rb repressor complexes. Our results revealed that endocycle tissues show a strong requirement for the E2F–Rb active repression complexes during development. We demonstrated that an important role for the E2F–Rb complexes in endocycling cells is the downregulation of cyclin E during the gap phase.
Cyclin E is an important regulator of endocycle cells. E2F transcription factors are generally found to be important for the expression of cyclin E and for S phase entry or progression of the endocycle cells (Duronio et al., 1995; Royzman et al., 1997). How can removal of active repression in combination with an activating E2F mutation lead to decreased endoreplication? One possibility is based on the hypothesis that cyclin E will downregulate itself once its activity reaches a certain threshold (Lilly and Spradling, 1996). If cyclin E oscillation is completely dependent upon E2F transcriptional activation and cyclin E activity, one might predict that in de2f2;dE2F1su89 double mutants, unregulated dE2F1su89 might lead to higher cyclin E levels earlier than normal and thus trigger earlier cyclin E downregulation, which will result in less DNA replication (particularly a decreased heterochromatic sequence replication) during each round of the endocycle. A second possibility is that the length of each round of endocycle is increased, leading to a decreased number of endocycles during development. We found that salivary gland cells in de2f2;dE2F1su89 mutants show a normal representation of heterochromatic sequences (Figure 4K) but less overall DNA content (Figure 4L–O). These results suggest that the amount of DNA replicated during each round of the endocycle is not affected, but the number of endocycles is decreased in de2f2;dE2F1su89 mutants.
The decreased number of endocycles could be due to a lengthening of the S phase or a lengthening of the gap phase. Lengthening of the S phase would lead to an increased number of cells that are in the S phase, while lengthening of the gap phase would decrease the number of cells that are in S phase at any given time. A decreased number of S phase nuclei was observed in de2f2;dE2F1su89 salivary glands compared with that in wild-type salivary glands. Thus the gap phase of the endocycles in the de2f2;dE2F1su89 mutants was significantly lengthened. We found that de2f2;dE2F1su89 but not wild-type salivary gland cells accumulate high levels of cyclin E in some gap phase cells (cells that were not incorporating BrdU, see Figure 5M–P, arrow). As downregulation of cyclin E levels is required for continuous endoreplication (Follette et al., 1998; Weiss et al., 1998), the failure to downregulate cyclin E levels properly in these gap phase cells would probably inhibit endoreplication and lead to severe defects in tissues that require extensive endoreplication during development. The observation that decreasing the gene dosage of cyclin E partially suppressed the de2f2;dE2F1su89 phenotypes such as salivary gland endoreplication defects, macrochaetae defects and lethality provides strong support for the idea that the failure to downregulate cyclin E levels in these gap phase cells is a cause for the observed defects in de2f2;dE2F1su89 endocycle tissues.
Although previous results established that cyclin E oscillation is critical for continuous endoreplication (Follette et al., 1998; Weiss et al., 1998), it is not clear how cyclin E oscillation in endocycle cells is achieved. No cyclin E oscillation defect was observed in salivary gland cells in the dE2F1su89 mutants (data not shown), suggesting that active repression by the dE2F2–RBF complexes was sufficient to downregulate the level of cyclin E during the gap phase, even in the presence of the unregulated dE2F1su89. In contrast, removal of the dE2F2–RBF complexes in the dE2F1su89 background results in extensive defects in endocycle tissues and defective cyclin E downregulation in the gap phase of endocycling cells. These results argue strongly that the E2F–RBF complexes are required for the normal downregulation of cyclin E in the gap phase of endocycling cells. These results, in conjunction with the previous observation that E2F activity is required for cyclin E expression and S phase progression of endocycle cells, suggested a model in which E2F activation is required for S phase of the endocycles and active repression by E2F–Rb complexes is required during gap phase. It is interesting to note that even in the complete absence of RBF–E2F active repression, there are still significant levels of endoreplication, suggesting that the oscillation of cyclin E activity, although defective, can still occur to some extent in de2f2;dE2F1su89 mutants. It is possible that additional mechanisms such as protein degradation or binding to inhibitor proteins such as Dacapo can also contribute to the downregulation of cyclin E activity.
This model is also consistent with the observation that endocycle tissues but not mitotic tissues are severely affected in de2f2;dE2F1su89 mutants. As discussed above, in the absence of E2F–Rb repressor complexes, the downregulation of cyclin E is defective in the gap phase of endocycling cells, resulting in a block in endoreplication, growth and potentially terminal differentiation, and leading to severe developmental defects in the endocycle tissue. In contrast, it appears that mitotic cycles do not have a strict requirement for cyclin E oscillation. For example, it was shown previously that early embryonic division cycles proceed with constant cyclin E–cdc2c kinase activity (Sauer et al., 1995). In addition, studies of embryos that are devoid of maternal as well as zygotic RBF revealed that RBF was not required for these earlier mitotic cycles that do not have a G1 regulation (Du and Dyson, 1999). Thus while the cell cycle control in the mitotic tissues was not completely normal, lack of active repression and cyclin E oscillation in mitotic cells did not lead to a block in cell cycle progression, growth or differentiation, allowing those mitotic tissues to develop into relatively normal structures. The precise mechanism that underlies the different requirements for cyclin E oscillation between mitotic cycles and endocycles are not clear at present and will require further study.
Interestingly, the salivary glands from the de2f2; dE2F1su89 mutants showed defects not only in endoreplication and growth but also in the expression of genes such as lysozymes and trypsin/chymotrypsin-like proteases (data not shown). These genes are highly expressed in the wild-type salivary glands and are potentially important for salivary functions such as digestion of food. The failure to express these genes is an indication that the de2f2;dE2F1su89 salivary glands may have terminal differentiation defects. In addition, the de2f2;dE2F1su89 mutants also show a macrochaetae phenotype similar to the partially rescued rbf mutants (Du, 2000). While it is quite likely that the observed phenotypes are a consequence of the endoreplication defect, these terminal differentiation-related phenotypes associated with a lack of RBF function are likely to be relevant to some of the observations made in mice when the Rb–E2F proteins are altered. In particular, both uncontrolled E2F activity and loss of E2F activity were shown to cause defects in tissues with endocycles. For example, E2F-1 expression in megakaryocytes led to a block of terminal differentiation and accumulation of massive numbers of megakaryocytes (Guy et al., 1996). Furthermore, a recent study showed that knockout of DP1 resulted in a failure of extra-embryonic development, including a defect in endoreplication of the trophoblast giant cells (Kohn et al., 2003). In addition, it was shown recently that loss of Rb in the placenta leads to abnormal trophoblast stem cell proliferation or differentiation and that the embryonic lethality of Rb knockout mice can be largely attributed to a placental defect (Wu et al., 2003). Trophoblast giant cells are one cell lineage derived from trophoblast stem cells. As these giant cells become polyploid via endocycles, it will be interesting to examine whether a defect in endoreplication may also contribute to the defects observed in the Rb mutant placenta.
Materials and methods
Fly strains
The following fly strains were used in this study: dE2F1su89 and de2f2p111-5 (this study), l(2)k07215k07215 (Flybase), de2f276Q.1 (Frolov et al., 2001), GMRRBF4C, GMRRBF-2802, GMRRBFDap and UASRBF (Xin et al., 2002), cyclin EAR95 (Knoblich et al., 1994) and ptc (Duman-Scheel et al., 2002).
Lethality analysis of the de2f2;dE2F1su89 and cycE/+,de2f2;dE2F1su89 flies
For lethality analysis of the de2f2;dE2F1su89 flies, de2f276Q.1/CyO(GFP); dE2F1su89/Tm6B flies were crossed to de2f2p111-5/CyO(GFP);dE2F1su89/Tm6B flies. Non-GFP and non-Tb larvae of the second or early third instar were identified, and the development of these larvae was followed to determine the survival to the pupae and adult stages. The survival of >100 correct genotype larva was determined for each genotype, and Fisher’s exact test was used to determine the statistical significance.
Scanning electron microscopy, antibody staining and salivary gland DNA content quantification
Flies were dehydrated and mounted as previously described (Du, 2000). The antibody staining procedure was as described (Duman-Scheel et al., 2002). Salivary gland DNA content was determined essentially as described (Weiss et al., 1998). Briefly, salivary glands were dissected from wandering third instar larvae, fixed and labeled with 1 µg/ml DAPI at room temperature for 1 h. Images of salivary glands were captured using the Zeiss Axiocam with a fixed exposure time and analyzed by Adobe Photoshop. To quantify the DNA labeling intensities, the pixel intensities within nuclei of one focal plane including the nuclear equator were summed. For each genotype, >40 nuclei from five different salivary glands were examined. Student’s t-test was used to determine the statistical significance.
Biochemical analysis
Yeast two-hybrid assays and CAT assays were carried out as described (Xin et al., 2002). Sequencing was carried out using the dRhodamine terminator cycle sequencing kit from PE Applied Biosystems. Immunoprecipitation–western blotting was carried out as described (Du et al., 1996a). Determination of the heterochromatic sequence representation was carried out as described (Lilly and Spradling, 1996) using rDNA as the heterochromatic probe and the P1 clone DS07108 as the euchromatic probe. Mitotically active diploid disc DNA, which is considered to have 100% representation of all the heterochromatic sequences, was used for normalization. The percentages of heterochromatic sequence representations from four independent DNA samples were averaged and repeated. No difference in percentage heterochromatic sequence representation was observed between wild-type and the de2f2;dE2F1su89 flies.
BrdU incorporation/in situ hybridization
BrdU staining for eye discs was performed as previously described (Du, 2000). For the BrdU labeling of the pupal notum, staged pupae were collected and dissected in M3 medium and were placed in 1 mg/ml BrdU/M3 solution with gentle shaking for 1.5 h at room temperature for incorporation. For salivary gland BrdU staining, salivary glands were dissected from early third instar larvae and were incubated for 2 h in 1 mg/ml BrdU/M3 solution followed by treatment as described above. In situ hybridization was carried out as described (Du, 2000).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We would like to thank Dan Zhu, Yanfei Xu and Aadrianna Cravens for technical assistance at different stages of this work. We thank Dr G.Leone for communicating results prior to publication, Dr N.Dyson for the de2f2 mutant flies, Dr H.Richardson for the anti-cyclin E antibody, the Bloomington Drosophila Stock Center for fly stocks, Ed Williamson and the University of Chicago SEM facility for help with the SEM, Jenny Pogoriler for help during manuscript preparation, and Drs Kay Macleod and Chip Ferguson for critical reading of the manuscript. This work was supported in part by funds from the Cancer Research Foundation, the Sydney Kimmel Foundation for Cancer Research, and a research grant from NIH to W.D. W.D. is a Leukemia and Lymphoma Society Scholar.
References
- Brook A., Xie,J.-E., Du,W. and Dyson,N. (1996) Requirements for dE2F function in proliferating cells and in post-mitotic differentiating cells. EMBO J., 15, 3676–3683. [PMC free article] [PubMed] [Google Scholar]
- Cayirlioglu P., Bonnette,P.C., Dickson,M.R. and Duronio,R.J. (2001) Drosophila E2f2 promotes the conversion from genomic DNA replication to gene amplification in ovarian follicle cells. Development, 128, 5085–5098. [DOI] [PubMed] [Google Scholar]
- de Nooij J.C., Letendre,M.A. and Hariharan,I.K. (1996) A cyclin-dependent kinase inhibitor, Dacapo, is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell, 87, 1237–1247. [DOI] [PubMed] [Google Scholar]
- Du W. (2000) Suppression of the rbf null mutants by a de2f1 allele that lacks transactivation domain. Development, 127, 367–379. [DOI] [PubMed] [Google Scholar]
- Du W. and Dyson,N. (1999) The role of RBF in the introduction of G1 regulation during Drosophila embryogenesis. EMBO J., 18, 916–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W., Vidal,M., Xie,J.E. and Dyson,N. (1996a) RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes Dev., 10, 1206–1218. [DOI] [PubMed] [Google Scholar]
- Du W., Xie,J.-E. and Dyson,N. (1996b) Ectopic expression of dE2F and dDP induces cell proliferation and death in the Drosophila eye. EMBO J., 15, 3684–3692. [PMC free article] [PubMed] [Google Scholar]
- Duman-Scheel M., Weng,L., Xin,S. and Du,W. (2002) Hedgehog regulates cell growth and proliferation by inducing cyclin D and cyclin E. Nature, 417, 299–304. [DOI] [PubMed] [Google Scholar]
- Duronio R.J. and O’Farrell,P.H. (1995) Developmental control of the G1 to S transition in Drosophila; cyclin E is a limiting downstream target of E2F. Genes Dev., 9, 1456–1468. [DOI] [PubMed] [Google Scholar]
- Duronio R.J., O’Farrell,P.H., Xie,J.-E., Brook,A. and Dyson,N. (1995) The transcription factor E2F is required for S phase during Drosophila embryogenesis. Genes Dev., 9, 1445–1455. [DOI] [PubMed] [Google Scholar]
- Duronio R.J., Brook,A., Dyson,N. and O’Farrell,P.H. (1996) E2F-induced S phase requires cyclin E. Genes Dev., 10, 2505–2513. [DOI] [PubMed] [Google Scholar]
- Dynlacht B.D., Brook,A., Dembski,M.S., Yenush,L. and Dyson,N. (1994) DNA-binding and trans-activation properties of Drosophila E2F and DP proteins. Proc. Natl Acad. Sci. USA, 91, 6359–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N. (1998) The regulation of E2F by pRB-family proteins. Genes Dev., 12, 2245–2262. [DOI] [PubMed] [Google Scholar]
- Edgar B.A. and Orr-Weaver,T.L. (2001) Endoreplication cell cycles: more for less. Cell, 105, 297–306. [DOI] [PubMed] [Google Scholar]
- Follette P.J., Duronio,R.J. and O’Farrell,P.H. (1998) Fluctuations in cyclin E levels are required for multiple rounds of endocycle S phase in Drosophila. Curr. Biol., 8, 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov M.V., Huen,D.S., Stevaux,O., Dimova,D., Balczarek-Strang,K., Elsdon,M. and Dyson,N.J. (2001) Functional antagonism between E2F family members. Genes Dev., 15, 2146–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubatz S., Lindeman,G.J., Ishida,S., Jakoi,L., Nevins,J.R., Livingston,D.M. and Rempel,R.E. (2000) E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol. Cell, 6, 729–735. [DOI] [PubMed] [Google Scholar]
- Guy C.T., Zhou,W., Kaufman,S. and Robinson,M.O. (1996) E2F-1 blocks terminal differentiation and causes proliferation in transgenic megakaryocytes. Mol. Cell. Biol., 16, 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V. and Posakony,J.W. (1989) Development of adult sensilla on the wing and notum of Drosophila melanogaster. Development, 107, 389–405. [DOI] [PubMed] [Google Scholar]
- He S., Cook,B.L., Deverman,B.E., Weihe,U., Zhang,F., Prachand,V., Zheng,J. and Weintraub,S.J. (2000) E2F is required to prevent inappropriate S-phase entry of mammalian cells. Mol. Cell. Biol., 20, 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J.A., Sauer,K., Jones,L., Richardson,H., Saint,R. and Lehner,C.F. (1994) Cyclin E controls S-phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell, 77, 107–120. [DOI] [PubMed] [Google Scholar]
- Kohn M.J., Bronson,R.T., Harlow,E., Dyson,N.J. and Yamasaki,L. (2003) Dp1 is required for extra-embryonic development. Development, 130, 1295–1305. [DOI] [PubMed] [Google Scholar]
- Lilly M.A. and Spradling,A.C. (1996) The Drosophila endocycle is controlled by cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev., 10, 2514–2526. [DOI] [PubMed] [Google Scholar]
- Ohtani K. and Nevins,J.R. (1994) Functional properties of a Drosophila homolog of the E2F1 gene. Mol. Cell. Biol., 14, 1603–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posakony J.W. (1994) Nature versus nurture: asymmetric cell divisions in Drosophila bristle development. Cell, 76, 415–418. [DOI] [PubMed] [Google Scholar]
- Royzman I., Whittaker,A.J. and Orr-Weaver,T.L. (1997) Mutations in Drosophila DP and E2F distinguish G1–S progression from an associated transcriptional program. Genes Dev., 11, 1999–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudkin G.T. (1972) Replication in polytene chromosomes. Results Probl. Cell Differ., 4, 59–85. [DOI] [PubMed] [Google Scholar]
- Sauer K., Knoblich,J.A., Richardson,H. and Lehner,C.F. (1995) Distinct modes of cyclin E/cdc2c kinase regulation and S-phase control in mitotic and endoreduplication cycles of Drosophila embryogenesis. Genes Dev., 9, 1327–1339. [DOI] [PubMed] [Google Scholar]
- Sawado T., Yamaguchi,M., Nishimoto,Y., Ohno,K., Sakaguchi,K. and Matsukage,A. (1998) dE2F2, a novel E2F-family transcription factor in Drosophila melanogaster. Biochem. Biophys. Res. Commun., 251, 409–415. [DOI] [PubMed] [Google Scholar]
- Smith A.V. and Orr-Weaver,T.L. (1991) The regulation of the cell cycle during Drosophila embryogenesis: the transition to polyteny. Development, 112, 997–1008. [DOI] [PubMed] [Google Scholar]
- Stevaux O., Dimova,D., Frolov,M.V., Taylor-Harding,B., Morris,E. and Dyson,N. (2002) Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. EMBO J., 21, 4927–4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi J.M. and Lees,J.A. (2002) Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol., 3, 11–20. [DOI] [PubMed] [Google Scholar]
- Trimarchi J.M., Fairchild,B., Wen,J. and Lees,J.A. (2001) The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc. Natl Acad. Sci. USA, 98, 1519–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub S.J., Prater,C.A. and Dean,D.C. (1992) Retinoblastoma protein switches the E2F site from positive to negative element. Nature, 358, 259–261. [DOI] [PubMed] [Google Scholar]
- Weintraub S.J., Chow,K.N.B., Luo,R.X., Zhang,S.H., He,S. and Dean,D.C. (1995) Mechanism of active transcriptional repression by the retinoblastoma protein. Nature, 375, 812–815. [DOI] [PubMed] [Google Scholar]
- Weiss A., Herzig,A., Jacobs,H. and Lehner,C.F. (1998) Continuous cyclin E expression inhibits progression through endoreduplication cycles in Drosophila. Curr. Biol., 8, 239–242. [DOI] [PubMed] [Google Scholar]
- Wu L. et al. (2003) Extra-embryonic function of Rb is essential for embryonic development and viability. Nature, 421, 942–947. [DOI] [PubMed] [Google Scholar]
- Xin S., Weng,L., Xu,J. and Du,W. (2002) The role of RBF in developmentally regulated cell proliferation in the eye disc and in cyclin D/Cdk4 induced cellular growth. Development, 129, 1345–1356. [DOI] [PubMed] [Google Scholar]
- Zhang H.S., Postigo,A.A. and Dean,D.C. (1999) Active transcriptional repression by the Rb–E2F complex mediates G1 arrest triggered by p16INK4a, TGFβ and contact inhibition. Cell, 97, 53–61. [DOI] [PubMed] [Google Scholar]