Abstract
Aberrant mRNAs containing premature termination codons (PTC-mRNAs) are degraded by a conserved surveillance system, referred to as the nonsense- mediated decay (NMD) pathway. Although NMD is reported to operate on the decapping and 5′-to-3′ exonucleolytic decay of PTC-mRNAs without affecting deadenylation, a role for an opposite 3′-to-5′ decay pathway remains largely unexplored. In this study, we have characterized the 3′-to-5′ directed mRNA degradation in the yeast NMD pathway. PTC-mRNAs are stabilized in yeast cells lacking the components of 3′-to-5′ mRNA-decay machinery. The 3′-to-5′ directed degradation of PTC-mRNAs proceeds more rapidly than that of the PTC-free transcript, in a manner dependent on the cytoplasmic exosome and Upf proteins. Moreover, Upf1p, but not Upf2p, interacts physically with an N-terminal domain of Ski7p, although the interaction requires Upf2p. The efficiency of 3′-to-5′ directed degradation of PTC-mRNAs is impaired by overexpression of Ski7p N-domain fragments that contain a sequence of the Upf1p-interaction region. These data suggest that the activation of 3′-to-5′ directed NMD is mediated through the interaction between Upf1p and the Ski7p N domain.
Keywords: exosome/nonsense-mediated decay/Ski complex/Ski7p/Upf proteins
Introduction
Eukaryotic cells have evolved mechanisms to ensure the fidelity of gene expression, especially in mRNA-decay pathways by degrading aberrant transcripts. One well studied example is nonsense-mediated mRNA decay, referred to as NMD (reviewed in Czaplinski et al., 1999; Hilleren and Parker, 1999). NMD is an RNA surveillance system that detects and destroys rapidly aberrant mRNAs containing premature termination codons (PTCs) to produce proper full-length proteins. The NMD pathway has been observed to function in all eukaryotic systems examined so far and is particularly well studied in the budding yeast, Saccharomyces cerevisiae. mRNA is normally degraded from two directions of 5′-to-3′ and 3′-to-5′ in the cytoplasm (reviewed in Caponigro and Parker, 1996; Mitchell and Tollervey, 2000). Both pathways begin with shortening of the 3′-poly(A) tail of mRNA. In the 5′-to-3′ decay, which is the major mRNA-degradation pathway in yeast, the poly(A) shortening triggers the removal of the 5′-cap structure by a decapping complex, Dcp1p/Dcp2p, exposing the transcript body to an exonuclease, Xrn1p, for rapid 5′-to-3′ decay (Muhlrad and Parker, 1992; Decker and Parker, 1993; Hsu and Stevens, 1993; Muhlrad et al., 1994, 1995; Beelman et al., 1996). It has been proposed that NMD is distinguished from the normal mRNA decay in that the decapping and 5′-to-3′ exonucleolytic decay occur without deadenylation (Muhlrad and Parker, 1994). Although the minor 3′-to-5′ decay pathway depends on a complex of exoribonucleases called the exosome (Muhlrad et al., 1995; Jacobs Anderson and Parker, 1998), its role in NMD remains largely unexplored.
Three factors involved in NMD have been identified in budding yeast (Leeds et al., 1991; Cui et al., 1995; He and Jacobson, 1995; Lee and Culbertson, 1995). Mutations in the UPF1, UPF2 and UPF3 genes selectively stabilize mRNA containing PTCs without affecting the decay rate of normal mRNA. These genes have also been identified in Caenorhabditis elegans, mammalian cells and the fission yeast Schizosaccharomyces pombe, indicating that the roles of Upf proteins are conserved among all eukaryotes (Pulak and Anderson, 1993; Perlick et al., 1996; Lykke-Andersen et al., 2000; Mendell et al., 2000; Serin et al., 2001). Upf1p is a cytoplasmic ATP-dependent RNA helicase; mutations affecting its ATPase activity also impair NMD (Czaplinski et al., 1995; Weng et al., 1996a). Upf2p rich in acidic amino acid residues interacts with both Upf1p and Upf3p (He et al., 1997). Upf3p, which is a smaller protein highly rich in basic residues, contains nuclear import and export signals, and it has been shown to localize to the nucleus (Shirley et al., 1998). Recognition of a termination codon as premature requires a cis-acting downstream element (DSE), which is located at the 3′ end of PTCs (Peltz et al., 1993; Zhang et al., 1995). The RNA-binding protein Hrp1p has been demonstrated to interact with the PGK1 DSE to provide a mark for PTC (Gonzalez et al., 2000). Hrp1p interacts directly with Upf1p, suggesting a direct link between Upf1p and a signal of PTC. Cells harboring single or multiple deletions of UPF genes exhibit equivalent stabilization of the nonsense mRNA, indicating that the Upf proteins either function as a complex or are components of the same regulatory pathway. However, their precise mechanism remains obscure. In addition, Upf2p and Upf3p interact with one of the polypeptide release factors, eRF3, and Upf1p interacts with both eRF1 and eRF3. Thus, Upf deletion enhances nonsense suppression, suggesting that yeast Upf proteins function in the translation termination process as well as in NMD (Czaplinski et al., 1998; Maderazo et al., 2000; Wang et al., 2001).
Degradation of 3′-to-5′ mRNA, both in the cytoplasm and in the nucleus, involves the exosome, which is highly conserved throughout evolution. The exosome is a complex of 3′-to-5′ exonucleases consisting of 10 subunits (Mitchell et al., 1997; Allmang et al., 1999). In the cytoplasm, the exosome participates in mRNA turnover (Jacobs Anderson and Parker, 1998; van Hoof et al., 2000). The function of the cytoplasmic complex requires several cofactors, named Ski proteins. Ski2p, a putative RNA helicase, Ski3p and Ski8p form a hetero-trimer referred to as the Ski complex, which is physically distinct from the exosome (Brown et al., 2000). The interaction between the exosome and the Ski complex is mediated through Ski7p, rather than by direct assembly (Araki et al., 2001). Ski7p is a GTP-binding protein consisting of two separated domains; one is the C-terminal region homologous to the GTP-binding elongation factor 1A (EF1A), and the other is an extra N-terminal domain that is not present in EF1A. We demonstrated previously that the N-terminal domain of Ski7p is required and sufficient, but its EF1A-like C domain is dispensable, for the 3′-to-5′ mRNA degradation (Araki et al., 2001). Furthermore, Ski7p is capable of interacting with both the exosome and the Ski complex physically through its N-terminal domain, and this interaction is required for the function of the cytoplasmic exosome. More recently, the C-terminal domain of Ski7p has been reported to be required for the non-stop decay pathway, which results in rapid decay of aberrant transcripts containing no stop codon (Frischmeyer et al., 2002; van Hoof et al., 2002).
In this study, we have characterized the 3′-to-5′ mRNA-decay pathway in yeast NMD and presented a role for exosome-mediated 3′-to-5′ mRNA decay. Interestingly, 3′-to-5′ directed degradation of mRNA containing PTCs proceeded more rapidly than that of the PTC-free transcript. Our present results suggest that the activation of 3′-to-5′ directed NMD is mediated through the interaction between Upf1p and Ski7p.
Results
Yeast strains lacking the components of 3′-to-5′ mRNA-decay machinery accumulate aberrant mRNA containing PTC
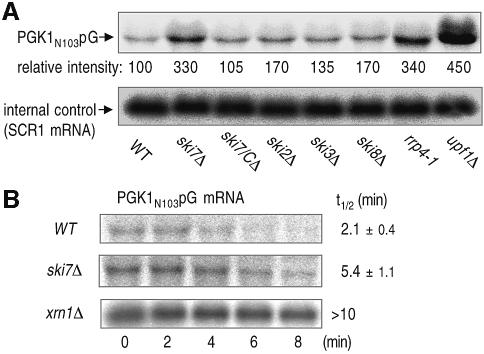
Although mRNA is normally degraded in both 5′-to-3′ and 3′-to-5′ directions after its deadenylation, it has been proposed that aberrant mRNA containing PTCs is subjected to decapping and 5′-to-3′ exonucleolytic decay without deadenylation (Muhlrad and Parker, 1994). To determine whether 3′-to-5′ mRNA decay is also involved in NMD, we first measured the steady-state level of a PGK1 transcript, PGK1N103pG, which contains a PTC at base 103 and a poly(G) tract in its 3′-untranslated region (Muhlrad and Parker, 1994; Muhlrad et al., 1995), in yeast strains that have defects in the components of cytoplasmic 3′-to-5′ mRNA-degradation machinery. The strains included in this study lack various SKI genes and we also used an rrp4-1 mutant which has a partial loss-of-function allele in exosome activity (Mitchell et al., 1996). These strains accumulated the PGK1N103pG transcript at different levels compared with the parental yeast strain (Figure 1A). Among them, a high level of accumulation was observed in the ski7Δ strain (second lane), and marked elevation of the mRNA was evident in upf1Δ (eighth lane) consistent with the previous report (Leeds et al., 1991). The inhibitory effect of ski7Δ on mRNA degradation appeared to be mediated through its N-terminal domain, since the deletion of ski7/C had no such effect (third lane). We further determined the actual decay rate of the nonsense transcript in ski7Δ. As shown in Figure 1B, the decay rate of PGK1N103pG mRNA in ski7Δ was significantly slower than that in the wild type, and much slower degradation of the mRNA was observed in xrn1Δ, which has a defect in the rapid 5′-to-3′ decay pathway. Thus, the 3′-to-5′ mRNA-decay pathway appeared to function in NMD, in addition to the previously identified 5′-to-3′ route.
Fig. 1. Yeast strains lacking the components of 3′-to-5′ mRNA-decay machinery accumulate an aberrant mRNA containing PTC, PGK1N103pG. (A) Yeast strains, wild type (WT, YS001), ski7Δ (YS002), ski7/CΔ (YS063), ski2Δ (YS060), ski3Δ (YS061), ski8Δ (YS014), rrp4-1 (YS094) and upf1Δ (YS065), which carry the PGK1N103pG reporter (pRP611), were grown at 26°C in a galactose-containing medium and harvested at mid-log phase. The aberrant mRNA was detected by northern blot analysis with end-labeled oligonucleotides specific for the PGK1N103pG (upper panel). The signals were quantified using a PhosphorImager and corrected for loading using the SRP RNA, scR1 (lower panel). The values under PGK1N103pG northern blot show relative intensity to the amount of mRNA obtained in the WT strain. (B) Yeast strains, wild type (WT, YS001), ski7Δ (YS002) and xrn1Δ (YS065), which carry the PGK1N103pG reporter (pRP611), were grown to mid-log phase in the galactose medium and shifted to a glucose medium to repress transcription of the reporter mRNA. Time points represent minutes after transcriptional repression. The half-lives (t1/2; min) of PGK1N103pG mRNA are shown as means ± standard deviations (SD), which were obtained from at least three independent experiments.
3′-to-5′ directed degradation is accelerated in mRNA containing PTC
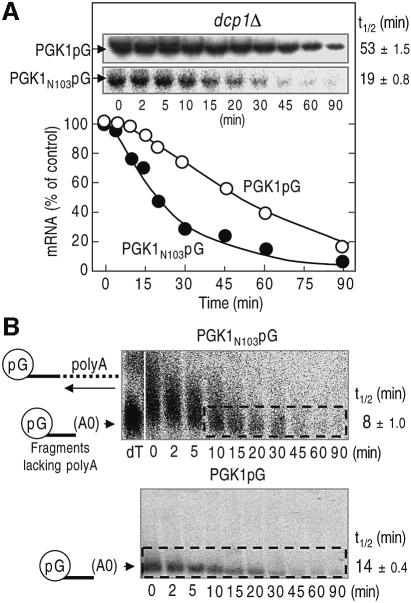
It has been reported that PTC increases the rate of 5′-to-3′ mRNA degradation. Therefore, we investigated whether the 3′-to-5′ mRNA-decay pathway is also accelerated in NMD. To estimate the contribution of the 3′-to-5′ decay pathway, we used a strain lacking the DCP1 gene, which has a defect in decapping and the following 5′-to-3′ decay. The reporter mRNAs, PGK1N103pG and the parental PGK1pG, used in the dcp1Δ mutant were under the control of the GAL1 UAS, and de novo induction was repressed in glucose medium to allow measurement of their decay rates. The PGK1N103pG mRNA was typified by a >2-fold increase in the rate of 3′-to-5′ directed degradation compared with the parental PGK1pG transcript (Figure 2A). We also observed that another NMD-susceptible transcript, the CPA1 mRNA, had the same behavior as the PGK1N103pG mRNA (Ruiz-Echevarria and Peltz, 2000; see Supplementary figure 1A available at The EMBO Journal Online).
Fig. 2. 3′-to-5′ mRNA degradation is accelerated in the aberrant mRNA containing PTC. (A) Yeast dcp1Δ strain (YS055) carrying PGK1pG (top panel and open circles) or PGK1N103pG (bottom panel and closed circles) reporters was grown to mid-log phase in the galactose medium and shifted to a glucose medium to repress transcription of the reporter mRNAs. After incubation for the indicated times, northern blot analysis was performed as described in Figure 1, and the amounts of transcript are illustrated as the function of the times. (B) Wild-type strain (YS001) that carries PGK1 N103pG (top panel) or PGK1pG (bottom panel) reporter was grown in a minimal medium containing galactose and shifted to the medium supplemented with glucose and cycloheximide to inhibit the transcription and decapping. After incubation for the indicated times, northern blot analysis was performed as described in Figure 1. The sample from the 0-min time that had been cleaved by RNaseH with oligo dT was also applied to the upper left lane (dT), which indicates the position of poly(G)–3′-end fragments lacking poly(A). The half-lives (t1/2; min) of mRNAs [full-length forms and fragments lacking poly(A) tail for A and B, respectively] are shown as means ± SD, which were obtained from three independent experiments.
It has been previously shown that when 5′-to-3′ decay is blocked by xrn1Δ, the nonsense transcripts are subject to deadenylation followed by 3′-to-5′ decay (Muhlrad and Parker, 1994). Therefore, the decay rate is controlled by both deadenylation and 3′-to-5′ mRNA decay. To prove that NMD triggers faster 3′-to-5′ decay, we directly measured the rate of 3′-to-5′ decay after deadenylation. Since the poly(G) tract forms a stable structure that inhibits 5′-exoribonuclease activity (Decker and Parker, 1993), 5′-to-3′ degradation of the mRNA produces an intermediate that stretches from the poly(G) tract to the 3′ end of the mRNA. The resultant intermediate is normally degraded by the 3′-to-5′ mRNA-degradation pathway (Jacobs Anderson and Parker, 1998). Consistent with the previous work (Muhlrad and Parker, 1994), the poly(G) to 3′-end fragments of the PGK1N103pG mRNA possess poly(A) tails (Figure 2B). Thus, we could roughly determine the decay rates of deadenylated mRNA fragments (upper panel, from 10 min time). As expected, the nonsense transcript was degraded more rapidly than the PTC-free transcript in the 3′-to-5′ direction. These results suggested that NMD triggers accelerated 3′-to-5′ decay.
3′-to-5′ directed NMD is mediated through the recognition of PTC by the releasing factor eRF3
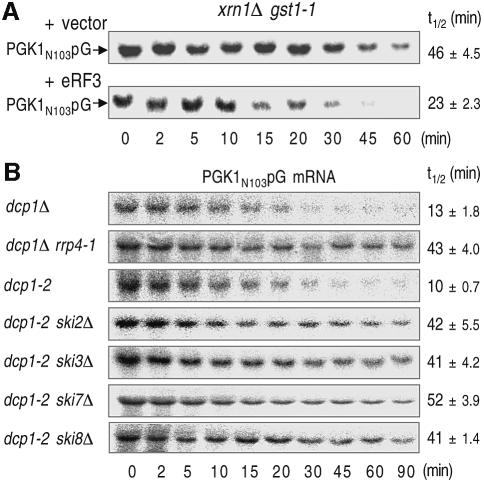
Additional experiments were performed to assess the role of recognition of a termination codon. We used a strain carrying a lesion in the eRF3 gene, gst1-1 (Kikuchi et al., 1988). This mutation causes a temperature-sensitive phenotype and has been demonstrated to increase the rate of the translation read-through of nonsense transcripts at the restrictive temperature for growth (T.Kobayashi, S.Hoshino and T.Katada, unpublished data). In this analysis, we used the xrn1Δ strain to exclude the contribution of 5′-to-3′ decay. As shown in Figure 3A, the xrn1Δ gst1-1 double mutation inhibited the degradation of PGK1N103pG mRNA at the restrictive temperature, and introduction of a plasmid containing wild-type eRF3/GST1 gene restored rapid degradation of the reporter mRNA. These results indicated that degradation of mRNA containing PTC is accelerated in the 3′-to-5′ direction and that the recognition of a PTC by eRF3 is responsible for this acceleration.
Fig. 3. 3′-to-5′ directed NMD is mediated through the recognition of PTC and requires both the exosome and the Ski proteins. (A) Yeast xrn1Δ gst1-1 (YS106) strains that carry PGK1N103pG reporter and either YCplac22-GST1 (bottom panel) or the empty vector (top panel) were grown at 25°C in the galactose medium to mid-log phase and incubated at 37°C for 1.5 h. (B) Yeast strains lacking the DCP1 gene (dcp1Δ, YS055; dcp1Δ rrp4-1, YS095) or containing its temperature-sensitive allele (dcp1-2, YS088; dcp11-2 ski2Δ, YS089; dcp1-2 ski3Δ, YS090; dcp1-2 ski7Δ, YS091; dcp1-2 ski8Δ, YS092) which carry the PGK1N103pG reporter were grown at 25°C in the galactose medium to mid-log phase and incubated at 37°C for 1.5 h. Transcription was repressed by the addition of glucose. After incubation for the indicated times, northern blot analysis was performed as described in Figure 1. The half-lives (t1/2; min) of mRNAs are shown as means ± SD, which were obtained from at least three independent experiments.
The cytoplasmic exosome and Upf proteins are required for the acceleration of 3′-to-5′ decay in NMD
We next investigated whether the exosome and Upf proteins function in the 3′-to-5′ NMD pathway. Loss-of-function mutations in proteins that are required for 3′-to-5′ mRNA degradation are synthetically lethal with lesions that block the other major decay pathway of decapping and 5′-to-3′ degradation (Jacobs Anderson and Parker, 1998). Therefore, we used a strain containing either dcp1-2 or rrp4-1 (Mitchell et al., 1996; Tharun and Parker, 1999). These are temperature-sensitive alleles and mRNA decay in these strains is severely inhibited at the restrictive temperature. To measure mRNA decay rates, these strains were grown at the permissive temperature and shifted to a restrictive temperature for 1.5 h. The dcp1Δ rrp4-1 double mutation increased the stability of PGK1N103pG transcripts at the non-permissive temperature compared with dcp1Δ (Figure 3B).
In addition to the cytoplasmic exosome, Ski2p, Ski3p, Ski7p and Ski8p are required for 3′-to-5′ decay of normal mRNA. Therefore, we also assayed the PGK1N103pG transcript in the following double mutants: dcp1-2 ski2Δ, dcp1-2 ski3Δ, dcp1-2 ski7Δ and dcp1-2 ski8Δ. These double-mutant strains allowed us to examine whether ski2Δ, ski3Δ, ski7Δ and ski8Δ alleles affect the 3′-to-5′ decay of the PGK1N103pG mRNA by comparison with the control dcp1-2 strain. In the dcp1-2 strain, mRNA degradation at the restrictive temperature occurs exclusively through the 3′-to-5′ pathway. As shown in Figure 3B, the PGK1N103pG transcript was stabilized in all of the double-mutant strains compared with dcp1Δ. These results implied that the 3′-to-5′ directed NMD requires both the exosome and the Ski proteins.
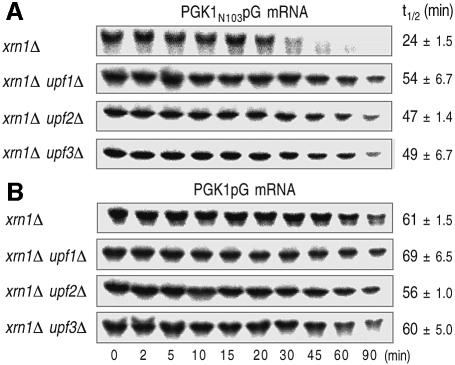
At least three trans-acting factors, Upf1p, Upf2p and Upf3p, have been observed to be essential for rapid degradation of mRNA containing PTCs in the 5′-to-3′ direction. The xrn1Δ upf1Δ, xrn1Δ upf2Δ and xrn1Δ upf3Δ double-mutant strains allowed us to examine whether the deletion of UPF genes affects the 3′-to-5′ decay in NMD by comparing with the control xrn1Δ strain. Deletion of any UPF genes in combination with xrn1Δ did not affect the stability of normal PGK1pG mRNA (Figure 4B), but caused significantly the stabilization of aberrant PGK1N103pG mRNA (Figure 4A). This observation suggested that Upf proteins are involved in the acceleration of 3′-to-5′ directed NMD.
Fig. 4. Acceleration of 3′-to-5′ directed NMD is mediated through Upf proteins. Yeast strains lacking XRN1 and/or UPF genes (xrn1Δ, YS064; xrn1Δ upf1Δ, YS070; xrn1Δ upf2Δ YS071; xrn1Δ upf3Δ, YS072), which carry PGK1N103pG (A) and PGK1pG (B) reporters, were grown at 25°C in the galactose medium to mid-log phase. Transcriptional repression and hybridization were performed as described for Figure 1. The half-lives (t1/2; min) of mRNAs are shown as means ± SD, which were obtained from at least three independent experiments.
Upf1p interacts physically with the N domain of Ski7p in a manner dependent on Upf2p
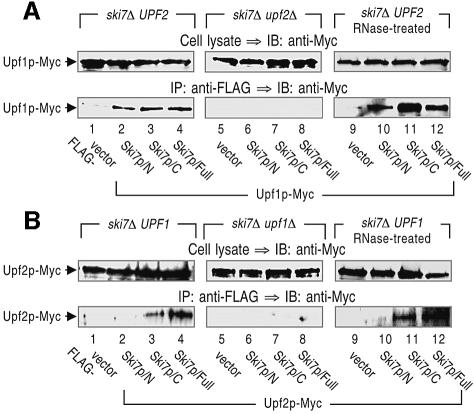
Based on the above results, we hypothesized that Upf proteins enhance 3′-to-5′ directed NMD by interacting with the N-terminal domain of Ski7p. This suggests a mechanism by which aberrant mRNA is targeted for degradation by the exosome. First, the findings in Figures 1 and 3B that the ski7Δ strain exhibits a significantly higher accumulation of the aberrant PGK1N103pG transcript than other mutant strains lacking the components of the Ski complex, namely, the SKI2, SKI3 and SKI8 genes, suggest that Ski7p especially plays a central role in the 3′-to-5′ directed NMD. Secondly, in contrast to the N-terminal deletion or the complete deletion of the SKI7 gene, the C-terminal truncation of Ski7p has no apparent effect on the accumulation of aberrant PGK1N103pG transcript (see Figure 1). Therefore, we investigated whether Ski7p is capable of interacting with Upf proteins.
For the analysis, the chromosomal copy of UPF1 or UPF2 was tagged with Myc epitope, and vectors expressing Ski7p variants fused with a FLAG epitope were introduced into the ski7Δ strain. Extracts were prepared from these strains and subjected to immunoprecipitation assay with an anti-FLAG antibody. Upf1p (Figure 5A, lane 2), but not Upf2p (Figure 5B, lane 2), was detected in the fractions precipitated with Ski7p/N (the amino acid sequence 1–264), indicating that Upf1p specifically interacts with the N-terminal domain of Ski7p. The interaction between Ski7p/N and Upf1p was independent of the existence of RNA, since pretreatment of the extracts with RNase A did not alter the immunoprecipitation profiles (Figure 5A, lanes 9–12). We further examined whether the deletion of the UPF2 gene could exert its influence on the Ski7p–Upf1p interaction. The association of Ski7p/N with Upf1p observed in the Upf2p-expressing strain was completely abolished in the upf2Δ strain (Figure 5A, lanes 5–8). These results argued that the interaction between Ski7p/N and Upf1p is dependent on the presence of Upf2p (see Discussion). Although the C-terminal domain of Ski7p (the sequence 265–747) is not required for the 3′-to-5′ directed NMD (see Figure 1), both Upf1p and Upf2p immunoprecipitated Ski7p/C (Figure 5A and B, lanes 3 and 11). The physiological significance of this interaction is currently under investigation.
Fig. 5. The N-terminal domain of Ski7p associates with Upf1p in a manner dependent on Upf2p. Cell extracts were prepared from SKI7- and/or UPF1/2-deletion strains that carry Upf1p-Myc (A; lanes 1–4, 9–12 for YS037–YS040 and lanes 5–8 for YS045–YS048) or Upf2p-Myc (B; lanes 1–4, 9–12 for YS041–YS044 and lanes 5–8 for YS049–YS052). The yeast strains also expressed the indicated forms of FLAG-tagged Ski7p. The extracts (lanes 1–8) were directly subjected to immunoprecipitation (IP) with an anti-FLAG. In some experiments, the extracts that had been treated with RNase were also subjected to the immunoprecipitation assay (lanes 9–12, RNase-treated). The precipitated proteins (bottom panels), together with the whole-cell extracts (top panels), were immunoblotted (IB) with an anti-Myc (9E10) antibody to detect Upf1p-Myc and Upf2p-Myc.
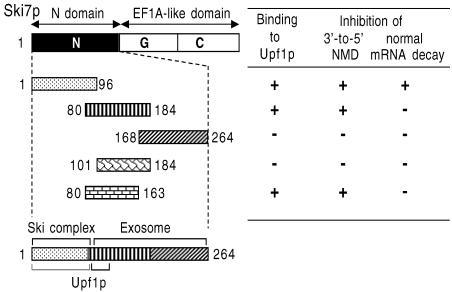
Since the N-terminal domain of Ski7p appeared to interact with Upf1p, we next mapped Upf1p-binding regions on the N domain. For this analysis, yeast strains expressing four different FLAG-tagged variants of Ski7p/N were generated, and their association with Upf1p was investigated by immunoprecipitation assay with the anti-FLAG antibody. We found that either of the two sequences of Ski7p, 1–96 or 80–184, was sufficient for the association with Upf1p (Figure 6A). Taken together, these results indicate that different regions on Ski7p/N are involved in the interaction with Upf1p.
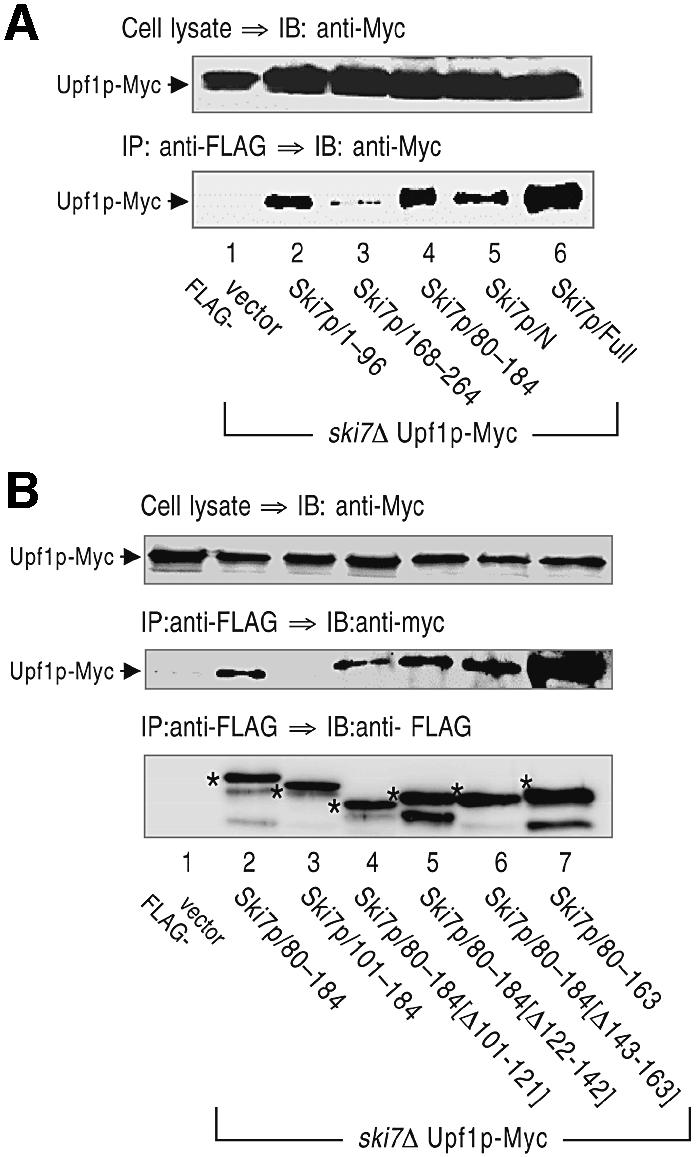
Fig. 6. Mapping of Upf1p-binding sites on the N-terminal domain of Ski7p. Cell extracts were prepared from yeast strains expressing Upf1p-Myc and the indicated forms of FLAG-tagged Ski7p (A, lanes 1–6 for YS037, YS101, YS103, YS104, YS038, YS039; and B, lanes 1–7 for YS037, YS038, YS123, YS124, YS125, YS126, YS127). The extracts were immunoprecipitated (IP) with the anti-FLAG antibody. The precipitated proteins (bottom panel) and whole-cell extracts (top panel) were immunoblotted (IB) with the anti-Myc antibody to detect Upf1p-Myc. The asterisks in (B) indicate FLAG-tagged Ski7p mutants expressed in the various strains.
Interaction of Ski7p/N with Upf1p is required for the 3′-to-5′ directed NMD
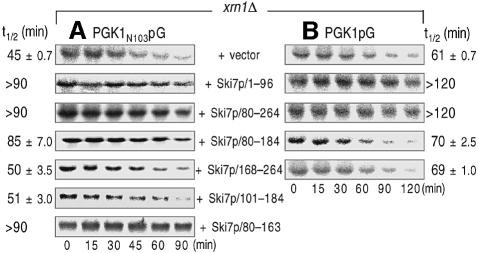
The above results are consistent with the hypothesis that Upf1p functions in the 3′-to-5′ exosome-mediated NMD through its interaction with Ski7p. In this model, it is predicted that interfering with the interaction between Ski7p/N and Upf1p would stabilize the 3′-to-5′ degradation of aberrant mRNAs containing PTCs. To test this prediction, we examined the degradation of the PGK1N103pG mRNA in the xrn1Δ strains overexpressing the deletion variants of Ski7p/N. It was found that three Ski7p/N mutants, namely the sequence 1–96, 80–264 and 80–184, induced significant inhibition of the 3′-to-5′ directed NMD (Figure 7A). Although there were some differences in the decay rate of the PGK1N103pG mRNA between xrn1Δ strains used in Figures 4 and 7, this may be due to the presence of the control vector, pTRPGAL1-His6-FLAG. However, we could examine whether overexpression of the Ski7p/N derivatives affects the 3′-to-5′ decay of the PGK1N103pG mRNA by comparing with the effect of vector alone. We previously showed that the variants of Ski7p N-terminal domain except the sequence 80–184 inhibit the normal 3′-to-5′ decay of another mRNA, MFA2pG, due to inhibition of the interaction of Ski7p with the Ski complex and the exosome (Araki et al., 2001; and see Discussion). Therefore, these results were not only consistent with the conclusion drawn from Figure 3B that the 3′-to-5′ NMD requires both the exosome and the Ski proteins, but also suggested that the 3′-to-5′ NMD is impaired by overexpression of Ski7p/80–184. The role of the 80–184 domain in NMD was further dissected with a series of deletion mutants. Immuno precipitation analyses identified the amino acid sequence 80–100 of Ski7p as an important region for the interaction with Upf1p (Figure 6B, lanes 3–6). Deletion of the Upf1p-interaction site (Ski7p/101–184) exerted no effect on the 3′-to-5′ directed NMD, although another fragment containing the 80–100 region (Ski7p/80–163) abrogated 3′-to-5′ directed NMD (Figure 7A). These results strongly suggested that 3′-to-5′ NMD requires the association between Upf1p and Ski7p.
Fig. 7. Interaction of the N-terminal domain of Ski7p with Upf1p is required for 3′-to-5′ directed NMD. The xrn1Δ strains expressing PGK1N103pG (A) and PGK1pG (B) reporters were transformed with plasmids carrying the indicated deletion mutants of Ski7p/N domain (vector, YS077; Ski7p/1–96, YS082; Ski7p/80–264, YS083; Ski7p/80–184, YS085; Ski7p/168–264, YS084; Ski7p/101–184, YS128; Ski7p/80–163; YS132). The transformants were grown in the galactose- containing medium and harvested at mid-log phase. Transcriptional repression and hybridization were performed as described for Figure 1. The half-lives (t1/2; min) of mRNAs are shown as means ± SD, which were obtained from at least three independent experiments.
Another likely mechanism is that the expression of Ski7p/80–184 might enhance the stability of PGK1N103pG mRNA independently of the presence of a nonsense codon. In order to explore this possibility, we overexpressed the deletion variants of Ski7p/N in the xrn1Δ strain expressing PGK1pG mRNA, which does not harbor any PTCs (Figure 7B). Consistent with our previous report, the normal 3′-to-5′ decay was impaired in yeast strains expressing the two deletion mutants, Ski7p/1–96 and Ski7p/80–264. However, the strain expressing Ski7p/80–184 exhibited no such defects. We also observed that Ski7p/80–184 reduced the 3′-to-5′ decay efficiency of another NMD-susceptible transcript, the CPA1 mRNA (see Supplementary figure 1B), although that of MFA2 mRNA (PTC-free transcript) was not impaired (data not shown). The different effects of Ski7p/80–184 observed between the normal and aberrant mRNAs further strengthened the above conclusion that the interaction of Ski7p/N with Upf1p is specifically important for the acceleration of 3′-to-5′ decay in NMD (Figure 8).
Fig. 8. The N domain of Ski7p interacting with Upf1p. Deletion mutants of Ski7p used in this study are shown, together with the summary of present results obtained in Figures 6 and 7.
Discussion
It has been proposed that NMD is distinguishable from the normal mRNA decay in that the decapping and 5′-to-3′ exonucleolytic decay occur without deadenylation. The role of the opposite 3′-to-5′ decay pathway in NMD, however, remains largely unexplored. In this study, we have characterized 3′-to-5′ directed mRNA decay in the yeast NMD pathway and presented a role for exosome-mediated 3′-to-5′ mRNA decay. We have found the following. (i) Yeast strains that have defects in the components of 3′-to-5′ mRNA-decay machinery accumulate the PGK1N103pG transcript harboring a PTC (Figure 1). (ii) The 3′-to-5′ directed mRNA decay is also accelerated in NMD, which requires the exosome, Ski proteins and Upf proteins (Figures 2–4; Supplementary figure 1A). (iii) Upf1p interacts with the N domain of Ski7p in a manner dependent on Upf2p but not on RNA (Figures 5 and 6), although it remains unknown whether the interaction is direct. (iv) Interaction of Upf1p with Ski7p/N appeared to be required for the acceleration of 3′-to-5′ mRNA decay (Figure 7; Supplementary figure 1B). We thus conclude that Upf1p mediates 3′-to-5′ mRNA degradation in NMD through its interaction with the N domain of Ski7p (Figure 8).
Previous work has suggested that 3′-to-5′ decay occurs slowly on nonsense mRNA and is observed only when the principal 5′-to-3′ degradation is inactivated (Muhlrad and Parker, 1994). In this study, we have explored the role of 3′-to-5′ decay in the yeast NMD pathway. Our results suggest that the 3′-to-5′ decay is also responsible for rapid degradation of nonsense transcripts. However, the contribution of the 3′-to-5′ pathway in nonsense mRNA decay is relatively lower than that of the 5′-to-3′ pathway. This is consistent with the fact that yeast predominantly use the 5′-to-3′ pathway for normal mRNA decay, and that the opposite 3′-to-5′ pathway is the minor event followed by deadenylation. Taken together, NMD in yeast can be viewed as a derivative of the major pathway. In contrast to the yeast system, mRNAs in mammalian cells are degraded primarily in the 3′-to-5′ direction by the cytoplasmic exosome (Chen et al., 2001; Wang and Kiledjian, 2001; Mukherjee et al., 2002). Therefore, it is tempting to speculate that the 3′-to-5′ pathway might have a prominent role in mammalian NMD.
Interestingly, only Upf1p was detected in Ski7p/N-precipitated fraction (Figure 5), although all three factors, Upf1p, Upf2p and Upf3p, function in 3′-to-5′ NMD (Figure 4). Our data argues that the Upf proteins participate in a more dynamic process. A number of observations support this possibility. (i) More than a single type of complex can be formed between the Upf proteins and release factors (Wang et al., 2001). (ii) Upf3p interacts directly with Upf2 protein, which in turn interacts directly with Upf1p, suggesting that Upf1p may be the last Upf protein to bind (Weng et al., 1996b; He et al., 1997). (iii) Association of Upf2p to polysomes requires the presence of Upf3p but not of Upf1p, whereas Upf1p and Upf3p are each found on polysomes independently of any other Upf proteins (Atkin et al., 1997). (iv) Upf1p is considerably more abundant than Upf2p or Upf3p: ∼1600, 160 and 80 molecules of Upf1p, Upf2p and Upf3p per yeast cell, respectively (Maderazo et al., 2000). Why the absence of Upf2p abolished the physical interaction between Ski7p/N and Upf1p remains to be elucidated (Figure 5A). While multiple scenarios can be envisaged, it is possible that a failure in surveillance complex assembly would impair proper folding or post-transcriptional modification of Upf1p. This possibility is consistent with previous findings that SMG-2, a homolog of Upf1p in C.elegans, is a phosphorylated protein and that SMG-3 (C.elegans Upf2p) is required for the phosphorylation of SMG-2 (Page et al., 1999).
We previously demonstrated that there are three regions on Ski7p/N, namely a Ski complex-binding region (the sequence 1–96) and two exosome-binding regions (the sequences 80–184 and 168–264). Overexpression of either the Ski complex-binding region or both exosome-binding regions inhibits 3′-to-5′ decay of the MFA2pG mRNA, suggesting dominant-negative effects, as might be expected when competition occurs for the binding of a critical protein that interacts with the N-terminus of Ski7p (Araki et al., 2001). In this study, an uncharacterized property of the sequence 80–184 of Ski7p was found to be functionally important for NMD and potentially forms a bridge between the NMD and the 3′-to-5′ decay machinery. This region is sufficient for the association with Upf1p (Figure 6). Overexpression of this small region resulted in significant inhibition of the 3′-to-5′ directed NMD, but deletion of the Upf1p-interaction site (Ski7p/101–184) exerted no effect on 3′-to-5′ directed NMD (Figure 7A; Supplementary figure 1A). It did not inhibit the normal 3′-to-5′ decay of the PGK1pG mRNA (Figure 7B) and the MFA2pG mRNA (data not shown). These observations indicate that the association between Ski7p and Upf1p is required for 3′-to-5′ directed NMD. Although the mechanism by which the interaction promotes decay of nonsense mRNAs deserves further investigation, we may propose simple models that are consistent with currently available results. There may be two ways in which the Ski7p–Upf1p complex can modulate the activity of the exosome so that the nonsense mRNAs are degraded rapidly. The Ski7p–Upf1p complex could stimulate their intrinsic enzyme activities, such as RNA helicase of Ski2p and exoribonucleases in the exosome. Alternatively, this complex could promote the association of the Ski complex with the exosome for the mRNA decay because Ski7p associates with the cytoplasmic form of the exosome and with the Ski complex through its N-terminal domain (Araki et al., 2001). These possibilities are not mutually exclusive.
Several observations suggest that Ski7p may be a critical component for the control of mRNA surveillance. First, Ski7p is required for the rapid degradation of nonsense mRNA in the 3′-to-5′ direction, and the ski7Δ strain exhibits a significantly higher accumulation of the aberrant PGK1N103pG transcript than other mutant strains lacking the components of the Ski complex (Figure 1). Secondly, the N-terminal domain of Ski7p interacts physically with Upf1p (Figures 5, 6 and 7). Thirdly, Ski7p has been also reported to play a principal role in the non-stop decay pathway, which results in rapid decay of aberrant transcripts containing no stop codon (van Hoof et al., 2002). In contrast to the non-stop decay, the EF1A-like domain of Ski7p is dispensable for 3′-to-5′ NMD (Figure 1), suggesting that mechanistic details of 3′-to-5′ NMD are different from those of the non-stop decay.
There are still many unresolved issues in the mechanism of the 3′-to-5′ directed NMD pathway. A subset of mutations of the UPF1 gene, which inactivated both its helicase and decay activities, still retained the ability to enhance translation termination at a nonsense codon and prevented nonsense suppression, and vice versa (Weng et al., 1996a,b). Therefore, it would be interesting to determine whether 3′-to-5′ NMD, 5′-to-3′ NMD and nonsense suppression are genetically separable functions of Upf1p. Furthermore, it would also be very interesting to determine whether NMD accelerates the deadenylation that precedes the decay of the mRNA body, and if so which deadenylase is responsible and how. These important issues are currently under investigation in our laboratory.
Materials and methods
Yeast strains and medium
All yeast strains used in this study are listed in Supplementary table I. The yeast cells were grown in standard culture media and transformed with DNA by the lithium acetate method. Disruption of yeast genes was achieved using PCR-based methods (Goldstein and McCusker, 1999; Araki et al., 2001), and the disruption was confirmed by the phenotypic analysis and/or PCR reactions with primers specific for the genes. Deletion of SKI7 and SKI8 genes was performed as described previously (Araki et al., 2001). The sequences of the oligonucleotides used are given in Table I.
Table I. Sequences of the oligonucleotides used for disruption of yeast genes.
| UPF1 | AATATACTTTTTATATTACATCAATCATTGTCATTATCAACCCTCGAGGTCGACGGTATC and GGTTTTTTAATTCCCCTTTAGCACCACGGCCAACCAAATTCGCTCTAGAACTAGTGGATC |
| UPF2 | TAATATTGTATCTGCATTGATAATACATTGGACAGAAATTCCCTCGAGGTCGACGGTATC and ATCATTTCACTGAAGAAGATGATGTTTTTAACGTTAATCTCGCTCTAGAACTAGTGGATC |
| UPF3 | GAGGGACTTACATTTCTGCTGAAATATATAGTAATCTATCCCCTCGAGGTCGACGGTATC and TGTATTCCATATATAATATATAAGAAGCCATGAGCTTTTACGCTCTAGAACTAGTGGATC |
| SKI2 | AACAACCTAACTCACAAAATTTACTGTACTAATACTAATTTATTCGAGGTCGACGGTATC and CTTTTATAAACATGACTCACATTGAGAATAAATGAGCTCTCGCTCTAGAACTAGTGGATC |
| SKI3 | GACACTAAGAACACAGAAAAGAAACACGAAGAGCAGAGGAAATTCGAGGTCGACGGTATC and GTTACATTAAGGTTTGATTGACTATCTCGAATCCAAATTTCGCTCTAGAACTAGTGGATC |
| XRN1 | AACACTTGTAACAACAGCAGCAACAAATATATATCAGTACGGTTCGAGGTCGACGGTATC and TAAAGTAACCTCGAATATACTTCGTTTTTAGTCGTATGTTCGCTCTAGAACTAGTGGATC |
| DCP1 | CATTTATCTTTGCAACACATCACAAGAAAAGCTGTGCACACACCGATCAACGTACAGTGG and TAAAAAAAATTCTCACTTGGGCATCTCACCTCTGTGCTCACCTGACAATCTGGCAGCTCG |
| RRP4 | TTGCATTCTAAATCTTTGCGTAGTCACACAAGCGTAAACAGATTCGAGGTCGACGGTATC and GTAGTTTATAAATGTGTAAACAGCGGCTTTTGTATTCCTACGCTCTAGAACTAGTGGATC |
| XRN1 for kan MX | ACTTGTAACAACAGCAGCAACAAATATATATCAGTACGGTCGGATCCCCGGGTTAATTAA and TAAAGTAACCTCGAATATACTTCGTTTTTAGTCGTATGTTGAATTCGAGCTCGTTTAAAC |
| UPF1 for hph MX | AATATACTTTTTATATTACATCAATCATTGTCATTATCAACGGATCCCCGGGTTAATTAA and AAGCCAAGTTTAACATTTTATTTTAACAGGGTTCACCGAAGAATTCGAGCTCGTTTAAAC |
Epitope tagging of UPF1, UPF2 and SKI7/N was performed by the one-step method described by Knop et al. (1999). Transformants were checked by PCR reactions for the correct integration. All epitope-tagged proteins expressed in this study were fully functional. The sequences of the oligonucleotides used are as follows: UPF1 (CAAAAGCATG AATTGTCAAAAGACTTCAGCAATTTGGGAATACGTACGCTGC AGGTCGAC and CAAGCCAAGTTTAACATTTTATTTTAACAGG GTTCACCGAAATCGATGAATTCGAGCTCG); UPF2 (CGAAACA AGATTAAAAAGATTGTTTTAAAACGTTCTTTCGACCGTACGC TGCAGGTCGAC and TATATAGTAAAAATTTAAAGAACTATA TTCGCAAAAGAATCATCGATGAATTCGAGCTCG); SKI7/N (CC TACTGAGTCGATTGATATTCATTCATTCATTGCCACCCATCG TACGCTGCAGGTCGAC and CAATAAGTATGAATGCCTAGTA TAATTTCTTAGTTGTAGGAATCGATGAATTCGAGCTCG).
Plasmid constructions
Plasmids for expressing the full-length and truncated forms of Ski7p were constructed as described previously (Araki et al., 2001). Plasmids harboring conditional dcp1-2 and rrp4-1 alleles were constructed as follows (Mitchell et al., 1996; Tharun and Parker, 1999). The wild-type genes were amplified from the genomic DNA by PCR using the following oligonucleotides: DCP1 (CCGGATCCTTCATATGATGACCGGAGC AGCAACTGC and GGGTCGACTCAAGCAAAAGAATCTTTTGGCT); RRP4 (CCGGATCCTTGAATTCATGTCCGAAGTTATCACAAT and GGGTCGACTTAGTTGCCGTTACCTCTCA). These fragments were subcloned into pBluescript II SK– vector. The conditional alleles were generated as described previously (Araki et al., 2001) with the following oligonucleotides (the mutation positions are underlined): dcp1-2 (GCATACGCTCCAAAAATAAAGCAACTACTT and CCCAATAACGTTGAAATTCAAAGCT); rrp4-1 (CAGTACCTATGTTGGGCTCT and CATGTTGCTTACCGCCAATG). DNA fragments of these alleles were subcloned into pTRPGAL1-His6-FLAG to generate pTRPGAL1-His6-FLAG-dcp1-2 and pTRPGAL1-His6-FLAG-rrp4-1 (Araki et al., 2001).
Immunoprecipitation of epitope-tagged proteins
Immunoprecipitation of FLAG-tagged proteins and subsequent detection of co-precipitated proteins were performed as follows. Logarithmically growing cells (5 × 108) in the selective medium containing 2% galactose were resuspended in 250 µl of a lysis buffer consisting of 50 mM Tris–HCl pH 7.4, 150 mM NaCl, 5 mM EDTA, 1 mM dithiothreitol, 0.1% NP-40 and protease inhibitors. The cells were mixed with glass beads (1 g) and disrupted by 10 cycles of vortexing for 30 s followed by incubating on ice for 1 min. The cell extracts were obtained by two consecutive runs of centrifugation (14 000 g for 10 min). After addition of anti-FLAG antibody-conjugated beads (M2-Agarose-AFFINITY; Sigma), the extracts (250 µl) were incubated on a rotator at 4°C for 2 h. The beads were pelleted and washed extensively with the lysis buffer without protease inhibitors. Proteins binding to the beads were eluted with an SDS–PAGE sample buffer by boiling for 5 min. In some experiments (Figure 6B), proteins were eluted with FLAG peptide (0.2 mg/ml) in the lysis buffer. The cell extracts (12.5 µl) and one-third of the eluted fraction were subjected to SDS–PAGE and immunoblotted with anti-Myc (9E10) and anti-FLAG (M2) monoclonal antibodies. To investigate whether the protein interaction was dependent on the existence of RNA, the lysates were also treated with RNase A (0.5 µg/µl) for 10 min at room temperature before the immunoprecipitation.
RNA analysis
Yeast cells were transformed by pRP602 and pRP611 plasmids encoding the PGK1pG and PGK1N103pG reporter mRNAs, respectively, under the control of the GAL1 promoter. Degradation of these reporters was detected as described previously (Muhlrad and Parker, 1994; Muhlrad et al., 1995). The strains containing dcp1-2, rrp4-1 or gst1-1 alleles were first grown in selective media containing 2% galactose at 25°C to mid-log phase (0.4–0.5 OD at 600 nm) and further incubated at 37°C. After 1.5 h, transcription of the reporters was blocked by washing once and addition of appropriate medium containing 4% glucose, and cells were further incubated at 37°C. Other strains were incubated at 25°C during all the experiments unless otherwise stated. At the indicated times, cells were removed, pelleted down and immediately frozen in liquid nitrogen. RNA that had been extracted by the hot-phenol procedure was separated by either 1% agarose gel or 4% polyacrylamide–7.5 M urea gel electrophoresis and transferred to the Hybond XL (Amersham Pharmacia). The reporters were detected by northern blotting using an oligonucleotide oRP121 (5′-AATTCCCCCCCCCCCCCCCCCCA-3′). The CPA1 transcript was detected as described by Ruiz-Echevarria and Peltz (2000). In each lane, the SRP RNA transcript, scR1, was used as an internal loading control by probing with an oligonucleotide o77 (5′-TCTAGCCGCGAGGAAGGA-3′).
All experiments were performed at least three times with different samples of the yeast strains, and the results were fully reproducible. Hence, most of the data shown are representative of several independent experiments.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to Drs Roy Parker, John McCusker and Richard Young for generous gifts of various plasmids and yeast strains. This work was supported in part by research grants from the ‘Research for the Future’ Program of the Japan Society for the Promotion of Science (JSPS-RFTF 96L00505), the Mitsubishi Foundation, and the Scientific Research Funds of the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government.
Note added in proof
While this manuscript was under revision, a part of our work was independently reported by Cao and Parker (Cell, 113, 533–545, 2003) and Mitchell and Tollervey (Mol. Cell, 11, 1405–1413, 2003).
References
- Allmang C., Petfalski,E., Podtelejnikov,A., Mann,M., Tollervey,D. and Mitchell,P. (1999) The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes Dev., 13, 2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y., Takahashi,S., Kobayashi,T., Kajiho,H., Hoshino,S. and Katada,T. (2001) Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J., 20, 4684–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin A.L., Schenkman,L.R., Eastham,M., Dahlseid,J.N., Lelivelt,M.J. and Culbertson,M.R. (1997) Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J. Biol. Chem., 272, 22163–22172. [DOI] [PubMed] [Google Scholar]
- Beelman C.A., Stevens,A., Caponigro,G., LaGrandeur,T.E., Hatfield,L., Fortner,D.M. and Parker,R. (1996) An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature, 382, 642–646. [DOI] [PubMed] [Google Scholar]
- Brown J.T., Bai,X. and Johnson,A.W. (2000) The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA, 6, 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G. and Parker,R. (1996) Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol. Rev., 60, 233–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y. et al. (2001) AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell, 107, 451–464. [DOI] [PubMed] [Google Scholar]
- Cui Y., Hagan,K.W., Zhang,S. and Peltz,S.W. (1995) Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev., 9, 423–436. [DOI] [PubMed] [Google Scholar]
- Czaplinski K., Weng,Y., Hagan,K.W. and Peltz,S.W. (1995) Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA, 1, 610–623. [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K., Ruiz-Echevarria,M.J., Paushkin,S.V., Han,X., Weng,Y., Perlick,H.A., Dietz,H.C., Ter-Avanesyan,M.D. and Peltz,S.W. (1998) The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev., 12, 1665–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K., Ruiz-Echevarria,M.J., Gonzalez,C.I. and Peltz,S.W. (1999) Should we kill the messenger? The role of the surveillance complex in translation termination and mRNA turnover. BioEssays, 21, 685–696. [DOI] [PubMed] [Google Scholar]
- Decker C.J. and Parker,R. (1993) A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev., 7, 1632–1643. [DOI] [PubMed] [Google Scholar]
- Frischmeyer P.A., van Hoof,A., O’Donnell,K., Guerrerio,A.L., Parker,R. and Dietz,H.C. (2002) An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science, 295, 2258–2261. [DOI] [PubMed] [Google Scholar]
- Goldstein A.L. and McCusker,J.H. (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast, 15, 1541–1553. [DOI] [PubMed] [Google Scholar]
- Gonzalez C.I., Ruiz-Echevarria,M.J., Vasudevan,S., Henry,M.F. and Peltz,S.W. (2000) The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol. Cell, 5, 489–499. [DOI] [PubMed] [Google Scholar]
- He F. and Jacobson,A. (1995) Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev., 9, 437–454. [DOI] [PubMed] [Google Scholar]
- He F., Brown,A.H. and Jacobson,A. (1997) Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol., 17, 1580–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P. and Parker,R. (1999) Mechanisms of mRNA surveillance in eukaryotes. Annu. Rev. Genet., 33, 229–260. [DOI] [PubMed] [Google Scholar]
- Hsu C.L. and Stevens,A. (1993) Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol., 13, 4826–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs Anderson A.R. and Parker,R. (1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J., 17, 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y., Shimatake,H. and Kikuchi,A. (1988) A yeast gene required for the G1-to-S transition encodes a protein containing an A-kinase target site and GTPase domain. EMBO J., 7, 1175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers,K., Pereira,G., Zachariae,W., Winsor,B., Nasmyth,K. and Schiebel,E. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast, 15, 963–972. [DOI] [PubMed] [Google Scholar]
- Lee B.S. and Culbertson,M.R. (1995) Identification of an additional gene required for eukaryotic nonsense mRNA turnover. Proc. Natl Acad. Sci. USA, 92, 10354–10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P., Peltz,S.W., Jacobson,A. and Culbertson,M.R. (1991) The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev., 5, 2303–2314. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu,M.D. and Steitz,J.A. (2000) Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell, 103, 1121–1131. [DOI] [PubMed] [Google Scholar]
- Maderazo A.B., He,F., Mangus,D.A. and Jacobson,A. (2000) Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol. Cell. Biol., 20, 4591–4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J.T., Medghalchi,S.M., Lake,R.G., Noensie,E.N. and Dietz,H.C. (2000) Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol. Cell. Biol., 20, 8944–8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. and Tollervey,D. (2000) mRNA stability in eukaryotes. Curr. Opin. Genet. Dev., 10, 193–198. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Petfalski,E. and Tollervey,D. (1996) The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev., 10, 502–513. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Petfalski,E., Shevchenko,A., Mann,M. and Tollervey,D. (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell, 91, 457–466. [DOI] [PubMed] [Google Scholar]
- Muhlrad D. and Parker,R. (1992) Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev., 6, 2100–2111. [DOI] [PubMed] [Google Scholar]
- Muhlrad D. and Parker,R. (1994) Premature translational termination triggers mRNA decapping. Nature, 370, 578–581. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker,C.J. and Parker,R. (1994) Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev., 8, 855–866. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker,C.J. and Parker,R. (1995) Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol., 15, 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee D., Gao,M., O’Connor,J.P., Raijmakers,R., Pruijn,G., Lutz,C.S. and Wilusz,J. (2002) The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J., 21, 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page M.F., Carr,B., Anders,K.R., Grimson,A. and Anderson,P. (1999) SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol., 19, 5943–5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz S.W., Brown,A.H. and Jacobson,A. (1993) mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev., 7, 1737–1754. [DOI] [PubMed] [Google Scholar]
- Perlick H.A., Medghalchi,S.M., Spencer,F.A., Kendzior,R.J.,Jr and Dietz,H.C. (1996) Mammalian orthologues of a yeast regulator of nonsense transcript stability. Proc. Natl Acad. Sci. USA, 93, 10928–10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulak R. and Anderson,P. (1993) mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev., 7, 1885–1897. [DOI] [PubMed] [Google Scholar]
- Ruiz-Echevarria M.J. and Peltz,S.W. (2000) The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell, 101, 741–751. [DOI] [PubMed] [Google Scholar]
- Serin G., Gersappe,A., Black,J.D., Aronoff,R. and Maquat,L.E. (2001) Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol. Cell. Biol., 21, 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley R.L., Lelivelt,M.J., Schenkman,L.R., Dahlseid,J.N. and Culbertson,M.R. (1998) A factor required for nonsense-mediated mRNA decay in yeast is exported from the nucleus to the cytoplasm by a nuclear export signal sequence. J. Cell Sci., 111, 3129–3143. [DOI] [PubMed] [Google Scholar]
- Tharun S. and Parker,R. (1999) Analysis of mutations in the yeast mRNA decapping enzyme. Genetics, 151, 1273–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A., Staples,R.R., Baker,R.E. and Parker,R. (2000) Function of the Ski4p (Csl4p) and Ski7p proteins in 3′-to-5′ degradation of mRNA. Mol. Cell. Biol., 20, 8230–8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A., Frischmeyer,P.A., Dietz,H.C. and Parker,R. (2002) Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science, 295, 2262–2264. [DOI] [PubMed] [Google Scholar]
- Wang W., Czaplinski,K., Rao,Y. and Peltz,S.W. (2001) The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J., 20, 880–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. and Kiledjian,M. (2001) Functional link between the mammalian exosome and mRNA decapping. Cell, 107, 751–762. [DOI] [PubMed] [Google Scholar]
- Weng Y., Czaplinski,K. and Peltz,S.W. (1996a) Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol. Cell. Biol., 16, 5477–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y., Czaplinski,K. and Peltz,S.W. (1996b) Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol. Cell. Biol., 16, 5491–5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Ruiz-Echevarria,M.J., Quan,Y. and Peltz,S.W. (1995) Identification and characterization of a sequence motif involved in nonsense-mediated mRNA decay. Mol. Cell. Biol., 15, 2231–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]