Abstract
Transformation of Mucor circinelloides with self-replicative plasmids containing a wild-type copy of the carotenogenic gene carB causes silencing of the carB function in 3% of transformants. Genomic analyses revealed a relationship between silenced phenotype and number of copies of plasmids. This phenotype results from a reduction of the steady-state levels of carB mRNA, a reduction that is not due to differences in the level of transcription, indicating that silencing is post-transcriptional. Small sense and antisense RNAs have been found to be associated with gene silencing in M.circinelloides. Two size classes of small antisense RNAs, differentially accumulated during the vegetative growth of silenced transformants, have been detected: a long 25-nucleotide RNA and a short 21-nucleotide RNA. Secondary sense and antisense RNAs corresponding to sequences of the endogenous gene downstream of the initial triggering molecule have also been detected, revealing the existence of spreading of RNA targeting in fungi. These findings, together with the self-replicative nature of the triggering molecules, make M.circinelloides a suitable organism for investigating some unresolved questions in RNA silencing.
Keywords: antisense RNA/post-transcriptional gene silencing/small interfering RNA/spreading of RNA targeting/transgene-induced RNA silencing
Introduction
RNA silencing, also termed post-transcriptional gene silencing (PTGS), involves sequence-specific RNA degradation and is triggered by the deliberate or fortuitous production of double-stranded RNA molecules (dsRNA) (for recent reviews see Chicas and Macino, 2001; Pickford et al., 2002; Tijsterman et al., 2002a; Cerutti, 2003). Introduction of transgenes, viral RNAs, transposons or dsRNA into a variety of hosts, ranging from protozoa to vertebrates, can induce PTGS. As a result, the expression of all endogenous genes sharing homology with the foreign nucleic acid is reduced or silenced through specific degradation of the mRNAs. Genetic dissection of PTGS, particularly in Neurospora crassa, Arabidopsis thaliana and Caenorhabditis elegans, has lead to the identification of several homologous genes required for gene silencing (Cogoni and Macino, 1997; Ketting and Plasterk, 2000; Mourrain et al., 2000). This supports the idea that the molecular basis of PTGS is similar in different organisms and suggests that PTGS is an ancestral gene regulation mechanism. Indeed, gene silencing seems to be a conserved mechanism that defends the genome against molecular parasites such as virus and transposons, and prevents the accumulation of abundant, non-functional messenger RNAs (Matzke et al., 2000).
Based on biochemical studies, a mechanism by which dsRNA triggers sequence-specific mRNA degradation has been proposed (reviewed in Cerutti, 2003). In this, a long dsRNA is processed into small interfering RNAs (siRNAs) of about 21–23 nucleotides (nt) in animals (Zamore et al., 2000) or 25 nt in plants and fungi (Hamilton and Baulcombe, 1999; Catalanotto et al., 2002), by the dsRNA-specific ribonuclease Dicer (Bernstein et al., 2001). Subsequently, the double-stranded siRNAs are incorporated into the multiprotein complex RISC (RNA-induced silencing complex) (Hammond et al., 2000), where the siRNAs are unwound. Antisense single-stranded siRNAs are used as a guide by the RISC complex to identify complementary RNAs, which are finally degraded by endonucleolitic and exonucleolitic cleavage. This pathway seems to be present in many of the eukaryotes where gene silencing has been described. However, how transgenes expressing sense mRNA are able to produce the dsRNA molecules that initiate the silencing mechanism is still an open question. Many models have been postulated, some of which invoke a nuclear step in transgene-induced gene silencing (reviewed in Chicas and Macino, 2001). The silencing state is transmitted between cells and through the syncytium of filamentous fungi, which has been termed systemic silencing (Cogoni et al., 1996; Voinnet and Baulcombe, 1997).
In the filamentous fungus Mucor circinelloides, transgenes are present as autonomous molecules, due to the self-replicative condition of the vectors used for transformation (Roncero et al., 1989), and can easily be recovered from the transformants (Navarro et al., 2000). This could represent an advantage in analysing molecular aspects of the silencing mechanism, since most of the transgenes used to induce silencing integrate into the genome of the host cell. The possibility that a gene silencing mechanism might be operating in M.circinelloides was postulated previously to explain the phenotype of transformants carrying additional wild-type copies of the regulatory gene crgA, a repressor of photocarotenogenesis (Navarro et al., 2001). Preliminary experiments carried out in our laboratory, using structural genes involved in the biosynthesis of carotenes, supported this hypothesis. These biosynthetic genes have been used in N.crassa as visual reporters of gene silencing, since any alteration of the carotenogenic pathway can be easily detected as a colour change in the colonies (Romano and Macino, 1992). In M.circinelloides, carotene biosynthesis is regulated by light (Navarro et al., 1995). Wild-type strain produces white-yellowish mycelium in the dark, which turns bright yellow after illumination with blue light due to the accumulation of β-carotene. Transcripts corresponding to the structural genes involved in the biosynthesis of carotenoids are barely detected in the dark, although their levels increase enormously in response to light (Velayos et al., 2000). Inactivation of the carB gene, which encodes the phytoene dehydrogenase enzyme, results in albino colonies due to the accumulation of the colourless precursor phytoene.
In this work we demonstrate the existence of a transgene-induced gene silencing mechanism in M.circinelloides, using the carB gene as a visual marker. We found that PTGS in M.circinelloides is associated with small sense and antisense RNAs, suggesting that the transcripts are converted to dsRNA and diced into siRNA molecules. Two size classes of antisense siRNA, differentially accumulated during vegetative growth, have been identified. Moreover, RNA targeting has been seen to spread from the initiator region into adjacent 3′ regions of the carB gene. To our knowledge, this is the first demonstration of spreading of RNA targeting in fungi. These results open up the way to investigate the nature of the signal that triggers transgene-induced PTGS and to study the roles of the two classes of siRNA in the induction, amplification and maintenance of gene silencing.
Results
Silencing of the carB gene
Transgene-mediated gene silencing in M.circinelloides was observed after transformation of a Leu– strain with a wild-type bright-yellow phenotype with self-replicative plasmids harbouring a Leu+ selective marker and different constructions of the carB gene (Figure 1A; Table I). About 3% of the transformants obtained with plasmid pMAT647, which contains the complete genomic carB sequence, showed an albino phenotype when incubated in the light, indicating alteration of the carotenoid biosynthetic pathway. The same behaviour was observed after introduction of plasmid pMAT754 containing the complete carB cDNA sequence controlled by the carB promoter. A functional transgene is not necessary to induce the albino phenotype, since 10% of transformants harbouring plasmid pMAT650, which contains a 3′-truncated carB gene, were unable to produce coloured carotenoids. The albino transformants had an unstable phenotype, as they segregated to wild-type or intermediate phenotypes after a cycle of vegetative growth on minimal medium (data not shown).
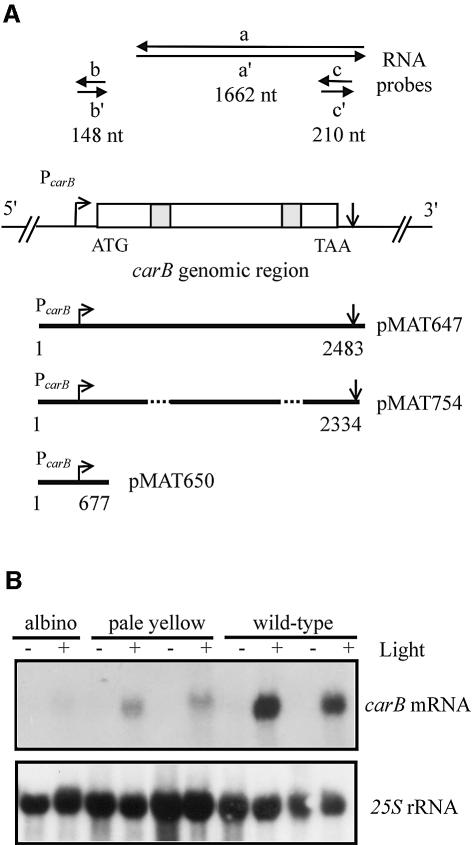
Fig. 1. (A) Schematic representation of the carB genomic region. The transcription start and polyadenylation sites (arrows), the translation start and termination codons, and the exons and introns (open and shaded boxes) are indicated. Above the scheme, carB riboprobes utilized to detect small sense and antisense RNAs. Below, carB constructs used for transformation. The length (in base pairs) of the carB sequence in each construct is indicated. (B) Northern blot analysis of transformants containing carB exogenous sequences. RNAs were extracted from dark-grown or light-pulsed mycelia of five pMAT754 transformants showing different phenotypes. About 5 µg of RNA was loaded in each lane and hybridized with a 1.8 kb cDNA fragment of the carB gene. The membrane was reprobed with a 25S rRNA probe to check loading. Illuminated mycelia were exposed to blue light for 4 min at 4 W/m2, and incubated in the dark for 20 min before RNA isolation. Densitometric analysis was used to estimate the mRNA levels.
Table I. Phenotypes of transformants obtained after the introduction of self-replicative plasmids containing different constructs of the carB gene into the wild-type strain.
| Plasmid | Transformants |
Total | Silencing frequency (%) | |
|---|---|---|---|---|
| Albino | Bright yellow | |||
| pMAT647 | 51 | 1561 | 1612 | 3.16 |
| pMAT754 | 12 | 373 | 385 | 3.12 |
| pMAT650 | 112 | 964 | 1076 | 10.4 |
| pLEU4 | 0 | 1758 | 1758 | 0 |
Plasmid pLEU4 was used as control. The colour of the transformants was observed after 48 h in the dark, followed by 24 h under illumination with blue light.
Gene silencing in M.circinelloides acts at the level of mRNA accumulation, as deduced from the northern blot analysis of the steady-state levels of carB mRNA in albino and wild-type transformants. Total RNAs were extracted from dark-grown and light-pulsed mycelia of transformants showing different degrees of the albino phenotype, and were hybridized with a carB probe (Figure 1B). Both albino and intermediate (pale yellow) transformants displayed barely detectable levels of carB mRNA in the light, as compared with the wild-type transformants. The carB mRNA level of the albino transformant was 18-fold lower than that of the wild-type ones, whereas intermediate phenotypes showed a 5-fold reduction in the carB transcript levels. These results indicate that differences in the severity of the albino phenotype are due to differences in the mRNA levels.
Genomic analysis of the silenced transformants
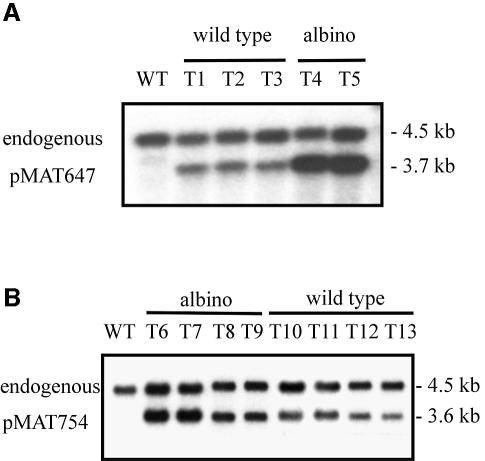
Genomic analysis of stable albino and wild-type transformants, selected after successive cycles of growth under selective pressure, indicated that the number of copies of plasmids in the silenced transformants was higher than in the wild-type ones. DNA from albino and wild-type transformants was digested with EcoRI and XhoI, and analysed by Southern blot hybridization (Figure 2). In all the transformants the endogenous carB (4.5 kb fragment) remained intact, as expected from the self-replicative nature of the recombinant plasmids. In addition, transformants contained an additional fragment that corresponds to exogenous carB sequences. Densitometric analysis revealed that albino transformants contained on average one to two molecules of plasmid per nucleus, whereas wild-type transformants contained <0.75 molecules per nucleus. Self-replicative plasmids in M.circinelloides tend to accumulate in a multimeric form (Navarro et al., 2001). Southern analysis showed that, in both classes of transformants, the plasmids seemed to be mainly organized as dimers and trimers, the sole difference between them being the total amount of plasmidic DNA (data not shown). This could eliminate gross rearrangement of the plasmid structure as the cause of silencing.
Fig. 2. Southern blot analysis of transformants containing carB exogenous sequences. Total DNA (0.75 µg) from the wild-type strain R7B (WT) and albino and wild-type transformants was digested with EcoRI and XhoI, and hybridized with a 1.8 kb cDNA fragment of carB. (A) Wild-type (T1-T3) and albino (T4-T5) transformants obtained with plasmid pMAT647. (B) Albino (T6-T9) and wild-type (T10-T13) transformants obtained with plasmid pMAT754. The EcoRI–XhoI digestion generates a 3.6 or 3.7 kb fragment from plasmids pMAT754 and pMAT647, respectively, which include the complete carB insert. The 4.5 kb fragment corresponds to the endogenous carB gene. Densitometric analysis of the hybridizing bands was used to calculate the copy number of carB exogenous sequences per nucleus.
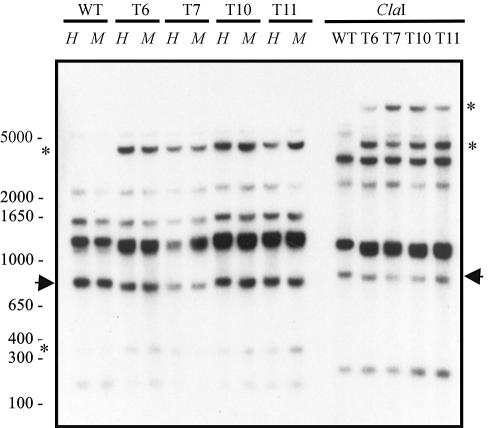
Methylation of the transgenic sequences has frequently been associated with silencing. In order to test the methylation state of the carB endogenous and exogenous sequences in the silenced transformants, DNA from albino and wild-type transformants were restricted with the pairs of isoschizomeric enzymes MspI and HpaII, and with the methylation-sensitive enzyme ClaI. Filters were hybridized with a probe that includes the carB coding sequence and promoter region. This probe can detect specific fragments derived from endogenous and exogenous carB sequences. No differences were observed between the pattern of the albino and wild-type transformants (Figure 3). In total, seven cytosines were analysed within the carB gene, two of them situated in the promoter region. The results indicate the absence of cytosine methylation at the sites tested and suggest that DNA methylation does not play a pivotal role in gene silencing in M.circinelloides.
Fig. 3. Analysis of the methylation state of carB exogenous and endogenous sequences in pMAT754 transformants. DNA of the wild-type stain R7B (WT) and albino (T6-T7) and wild-type (T10-T11) transformants was restricted with the isoschizomers MspI (M) and HpaII (H) (insensitive and sensitive to cytosine methylation, respectively), or with the enzyme ClaI, which is sensitive to cytosine methylation. Filter was hybridized with a 2.4 kb PstI–XhoI fragment of plasmid pMAT754, which contains the carB coding and promoter sequences. Arrows indicate fragments of 862 bp (HpaII–MspI digestion) and 888 bp (ClaI digestion), derived solely from endogenous carB sequences. Asterisks mark fragments exclusively derived from carB exogenous sequences. The sizes (in base pairs) of fragments of the 1 kb Plus ladder marker (Gibco-BRL) are indicated.
Gene silencing is post-transcriptional
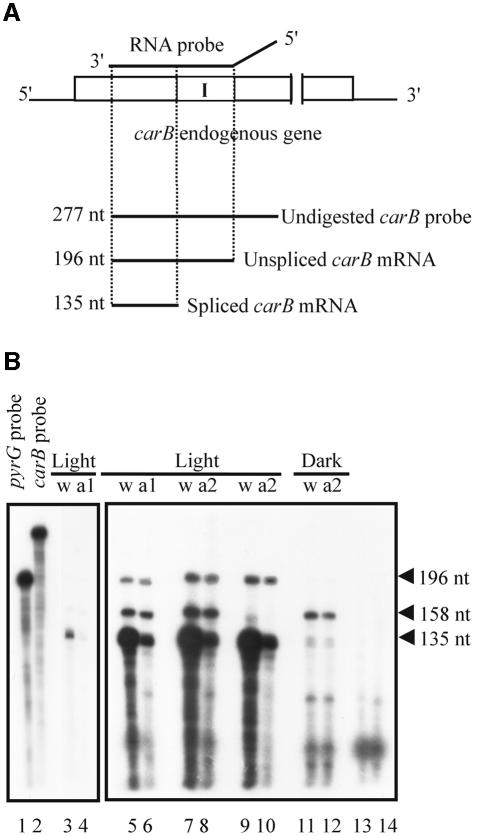
The reduction in the steady-state levels of carB mRNA detected in the albino transformants could be the result of a transcriptional or a post-transcriptional silencing mechanism. To distinguish between these two possibilities, RNase protection assays were performed on the wild-type strain and albino transformants. The relative amounts of the unspliced, primary transcript of carB and mature carB mRNA were determined by using a probe able to discriminate between them (Figure 4A). Since the carB expression is light inducible, mycelia were exposed to blue light for 4 min and incubated in the dark for 15 min before RNA was isolated. The results indicated that both albino and wild-type strains have the same level of unspliced carB mRNA precursor, while the amount of mature carB mRNA is roughly six times less in the silenced strains than in the wild-type (Figure 4B, lanes 5–8). To avoid possible contamination with the undigested pyrG probe, which was used as control to normalize the RNA quantity, RNase protection assays were also performed using only the carB probe (Figure 4B, lanes 9 and 10). Results confirm that the 196 nt fragment detected in these experiments corresponds to the unspliced carB mRNA. As expected, no signals corresponding to the precursor RNA were detected when RNA was isolated from dark-grown mycelia (Figure 4B, lanes 11 and 12). These results indicate that gene silencing in M.circinelloides occurs at a post-transcriptional level, affecting mature mRNA accumulation, but not the rate of transcription.
Fig. 4. RNase protection experiments for carB transcripts. (A) Schematic representation of endogenous carB transcripts, showing the expected sizes of fragments protected by unspliced and spliced carB mRNA. The rectangle represents the transcribed region of the endogenous carB gene. Shaded box indicates an intron (I) of the carB gene. The RNA probe (277 nt) was generated from plasmid pMAT643 and is indicated by a solid line above the carB gene. The probe contains sequences derived from the plasmid, which are indicated by the diagonal portion of the solid line. (B) RNase protection analysis was performed on total RNA isolated from light-pulsed or dark-grown mycelia of the wild-type strain (w) or two albino pMAT754 transformants (a1 and a2). A pyrG probe (202 nt) generated from plasmid pMAT645 was used as control to normalize the RNA quantity. This probe protects a 158 nt fragment of the pyrG mRNA. Lanes 1–4 correspond to a filter exposed for 1 h, and lanes 5–14 correspond to a filter exposed for 15 h. Lane 1, undigested pyrG probe; lane 2, undigested carB probe; lanes 3–8, RNase protection experiments using both the carB and the pyrG probes on RNA isolated from light-pulse mycelia; lanes 9 and 10, RNase protection experiments using only the carB probe; lanes 11 and 12, RNase protection experiments using both the carB and the pyrG probes on RNA isolated from dark-grown cultures; lane 13, carB probe digested with RNase; lane 14, pyrG probe digested with RNase.
Post-transcriptional gene silencing is associated with small antisense and sense RNAs
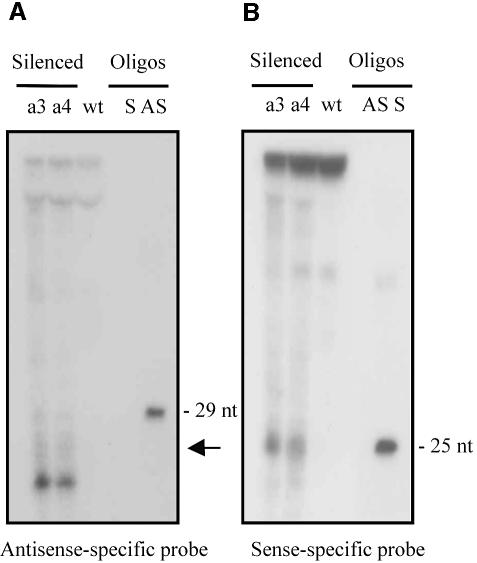
Transgene-mediated gene silencing of carB is associated with the presence of small RNAs corresponding to both sense and antisense strands (Figure 5). Those RNAs were identified by RNA blot hybridization of low-molecular-weight RNA samples isolated from the wild-type strain (unsilenced) or pMAT647 albino transformants (silenced), grown for 72 h in liquid culture. Identical RNA blots were hybridized with sense and antisense RNA probes able to recognize small RNAs derived from both transgenic and endogenous carB sequences (Figure 1A, probes a and a′). A predominant ∼21 nt antisense RNA was detected in the carB-silenced strains, whereas this RNA was absent from the wild-type strain (Figure 5A), meaning that post-transcriptional gene silencing in M.circinelloides is associated with the production of small antisense RNA molecules. Surprisingly, the size of the predominant small sense RNAs detected in the silenced strains was ∼25 nt (Figure 5B), in contrast to the results described in other eukaryotic systems, where both sense and antisense RNAs have a similar size. The same experiment was repeated using different samples of enriched low-molecular-weight RNAs isolated from different albino transformants. In all cases the disparity between the sense and antisense RNA sizes was confirmed (data not shown).
Fig. 5. Sense and antisense RNAs associated with post-transcriptional gene silencing of the carB gene. Fifteen micrograms of low molecular weight RNAs isolated from unsilenced wild-type strain (wt) or two different silenced albino transformants (a3 and a4) grown for three days under continuous illumination conditions were loaded in each lane. Ten picomoles per lane of 29-mer DNA oligonucleotide in antisense orientation (AS) and 25-mer DNA oligonucleotide in sense orientation (S) were used as size markers and to control the hybridization specificity. (A) RNA blot hybridized with a carB antisense-specific riboprobe (Figure 1A, probe a′). The same filter was re-hybridized with a carB sense-specific probe to detect the position of the 25 nt oligonucleotide (marked by an arrow). (B) Identical RNA blot hybridized with a carB sense-specific riboprobe (Figure 1A, probe a). Equal loading of the small RNA species was confirmed by EtBr staining of the predominant RNA species in the samples after the small RNAs were separated by agarose gel electrophoresis (data not shown).
Two classes of small antisense RNAs
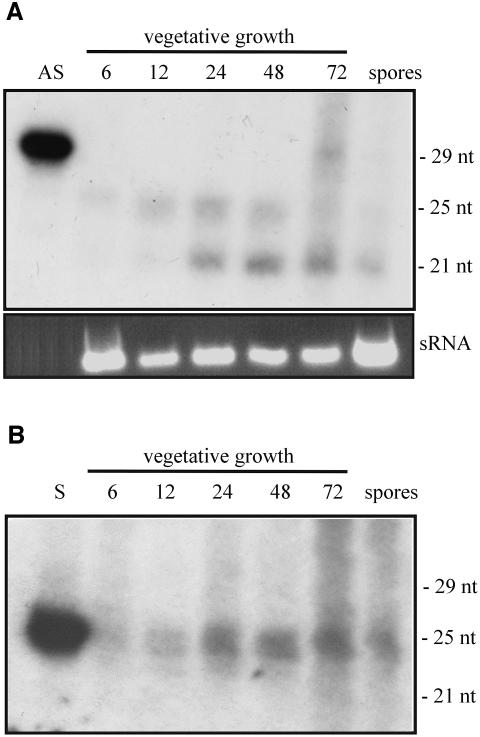
Post-transcriptional gene silencing in M.circinelloides is associated with two size classes of antisense RNA, which are differentially accumulated through the vegetative growth (Figure 6). Samples of enriched low-molecular-weight RNAs were isolated from albino strains after being grown for different times in liquid medium. RNA blot hybridization using the antisense-specific probe shows the presence of two size classes of antisense RNAs, 21 and 25 nt long (Figure 6A). The 25 nt RNAs are more abundant at the beginning of the growth cycle, whereas the 21 nt RNAs are prevalent after 48 h of growth. The same results were obtained when different small RNA samples isolated from different albino transformants were analysed (data not shown). The shorter antisense RNAs are transmitted through the spores, since they could be detected in small RNAs samples prepared from spores of albino transformants grown on solid medium. In contrast, no antisense RNAs were detected in the spores of the wild-type strain (data not shown). The accumulation of 21 nt RNAs in the spores is not responsible for the abundance of these species later during vegetative growth, since no spores are produced in liquid medium. When a similar blot was hybridized with the sense-specific probe (Figure 6B), only the 25 nt small RNAs were clearly detected in all the RNA samples. To rule out the possibility that the different size classes of antisense RNAs were an artefact due to possible contaminants in the RNA preparations, the electrophoretic mobility of labelled marker RNA was analysed alone or after mixing with the small RNAs obtained at 24 h. The same pattern of bands was observed in both samples (data not shown).
Fig. 6. Differential accumulation of 21 and 25 nt antisense RNAs during vegetative growth of silenced strains. Low-molecular weight RNAs were isolated from a silenced albino transformant of plasmid pMAT647 (a3), grown for different times (hours) in liquid culture and from spores collected from solid medium. Fifteen micrograms of RNA was loaded in each lane. Ten picomoles per lane of 29-mer DNA oligonucleotide in antisense orientation (AS) and 25-mer DNA oligonucleotide in sense orientation (S) were used as size markers. (A) RNA blot hybridized with a carB antisense-specific riboprobe (Figure 1A, probe a′). The predominant RNA species in the small RNA samples (sRNA) were stained with EtBr after RNAs were separated by agarose gel electrophoresis. (B) Identical RNA blot hybridized with a carB sense-specific riboprobe (Figure 1A, probe a).
Spreading of RNA targeting
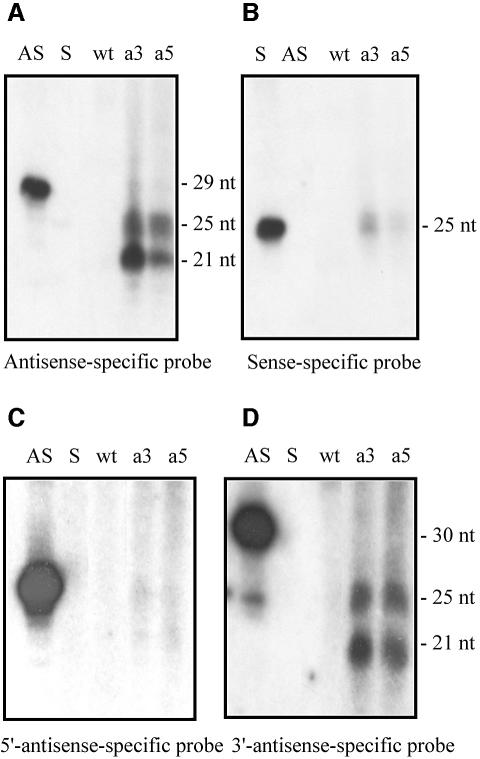
Small sense and antisense RNAs were also detected from sequences of the carB gene outside of the transgenic sequences (Figure 7). Low-molecular-weight RNA was isolated from albino transformants obtained with plasmid pMAT650, which contains a truncated version of the carB gene expanding from the promoter region to position 166 of the coding sequence. Those small RNAs were probed with sense and antisense-specific probes that do not detect exogenous carB sequences, but only adjacent downstream sequences derived from the endogenous gene (Figure 1A, probes a and a′). The two size classes of antisense RNA, 21 and 25 nt, were detected in the pMAT650 albino transformant (Figure 7A, a5), as was observed in albino transformants containing full-length carB exogenous sequences (Figure 7A, a3). When a similar blot was hybridized with the sense-specific probe, the 25 nt small RNAs were detected in both RNA samples (Figure 7B). Sense and antisense RNAs were absent in the wild-type strain. These results indicate that secondary sense and antisense RNAs are also produced from the targeted mRNA transcript, probably as derived products of mRNA degradation.
Fig. 7. Secondary sense and antisense RNAs in transgene-induced gene silencing. Low-molecular weight RNAs were isolated from the wild-type strain (wt), an albino transformant containing a full-length carB transgene (a3; plasmid pMAT647) and an albino transformant containing 3′-truncated carB exogenous sequences (a5; plasmid pMAT650). Cultures were grown for 24 h in liquid medium under continuous illumination conditions. Fifteen micrograms of RNA was loaded in each lane. (A) RNA blot hybridized with a carB antisense-specific riboprobe (Figure 1A, probe a′). (B) Identical RNA blot hybridized with the carB sense-specific riboprobe (Figure 1A, probe a). (C) RNA blot hybridized with the 5′-end carB antisense-specific riboprobe (Figure 1A, probe b′). (D) Identical blot hybridized with the 3′-end carB antisense-specific riboprobe (Figure 1A, probe c′). Ten picomoles per lane of 29-mer (A and B), 25-mer (C) or 30-mer (D) DNA oligonucleotides in antisense orientation (AS) and 25-mer DNA oligonucleotides in sense orientation (S) were used as size markers.
To determine whether the 25 and 21 nt antisense RNAs equally represent all portions of the target gene, the same RNA samples were hybridized to short antisense-specific probes corresponding to the 5′- and 3′-ends of the carB gene (Figure 1A, probes b′ and c′). Results showed that the two size classes of antisense RNAs were preferentially generated from the 3′ region of the carB gene, both in the transformants carrying a full-length transgene and in those harbouring a 3′-truncated transgene (Figure 7C and D). Similar blots probed with the 5′- and 3′-sense-specific riboprobes showed that the 25 nt sense RNAs were also produced from the 3′-end of the carB gene (data not shown).
Discussion
We used a simple visual reporter system to analyse transgene-induced gene silencing in M.circinelloides. Wild-type strains transformed with complete or truncated copies of the carB gene present an albino phenotype instead of the bright yellow colour seen when the carB gene is expressed at the wild-type levels. The albino phenotype is a consequence of strong reduction in the steady-state levels of carB mRNA. This reduction in the mature (spliced) mRNA is not due to differences in the rate of transcription, since levels of unspliced carB mRNA are the same in wild-type and albino transformants. This establishes that transgene-induced gene silencing in M.circinelloides occurs at a post-transcriptional level.
Only a small fraction (3%) of transformants containing a full-length carB transgene presents an albino phenotype. This frequency is much lower than that reported for the homologous gene al-1 in N.crassa (Romano and Macino, 1992). However, in M.circinelloides, unlike in N.crassa, transforming DNA does not integrate into the genome but is maintained in an episomal state. Thus, transgene expression is not affected by position effects or host regulatory sequences at insertion sites, both of which are thought to be involved in the production of abnormally processed RNAs. Those RNAs could be converted to dsRNA by the activity of an RNA-dependent RNA polymerase (RdRP) (Zamore, 2002). In this sense, it is important to note that the frequency of silencing increases when a plasmid containing a truncated version of the carB gene is used to transform the wild-type strain (Table I). Besides that, the low copy number of transgenes found in most of the transformants could also account for the low frequency of silencing, since our results reveal a relationship between silencing and copy number of transgenic sequences. A high concentration of abnormally processed RNA seems to be required to activate RdRP-mediated copying (Zamore, 2002; Tang et al., 2003), which would support this hypothesis. Alternatively, a low efficiency of the enzymes involved in the production of small RNAs could also explain the low efficiency of silencing. Even though no transgene rearrangements seem to occur in the silenced transformants, truncated or abnormally processed RNAs could be produced as a result of DNA–DNA pairing or epigenetic changes at the transgenic loci (Chicas and Macino, 2001).
Methylation of the coding region of transgenes has frequently been associated with post-transcriptional gene silencing (Cerutti, 2003). Here we failed to detect any cytosine methylation of either the exogenous or endogenous carB gene in silenced transformants. Recent evidence suggests that chromatin structure plays a part in the establishment of post-transcriptional gene silencing (Morel et al., 2000). Further, several reports have directly implicated the post-transcriptional silencing machinery in the methylation of histone H3 and heterochromatin formation at the target locus (Cerutti, 2003). The analysis of the histone methylation state in M.circinelloides silenced transformants will uncover whether chromatin modifications are components of the post-transcriptional silencing pathway in fungi.
Transgene-induced gene silencing is associated with small sense and antisense RNAs (siRNAs) in M.circinelloides, as has been described in other organisms (Tijsterman et al., 2002a). Surprisingly, two size classes of antisense RNAs are differentially accumulated during the vegetative growth of silenced transformants. Short (21 nt) antisense RNAs are primarily detected late during the vegetative growth, whereas a long (25 nt) antisense RNA is produced early in the vegetative life cycle. This long antisense RNA is almost undetectable in the spore RNA population, where the short 21 nt RNA is the predominant species. These results could indicate the existence of two classes of small siRNAs in M.circinelloides, the long 25 nt RNA and the short 21 nt RNA. The fact that we were unable to detect the 21 nt sense RNAs clearly, even upon longer exposure of the filters, could be used as an argument against the origin of the short antisense siRNAs from a dsRNA precursor. However, a similar situation has been described in wild-type C.elegans exposed to dsRNA in order to induce silencing, where only small RNA species of antisense polarity were detected (Tijsterman et al., 2002b). Recent work has demonstrated the existence of two size classes of siRNAs in plants (Hamilton et al., 2002). Long (24–26 nt) RNAs are correlated with systemic silencing and with the methylation of homologous DNA, whereas short (21–22 nt) RNAs are correlated with sequence-specific degradation. Using an in vitro system, Zamore and co-workers have demonstrated that different Dicer-like enzymes produce each class of siRNA in plants (Tang et al., 2003). It is tempting to speculate that a similar situation may take place in M.circinelloides. The long 25 nt RNA would be involved in the propagation of silencing through the coenocytic hyphae of the fungus and would mainly be produced during the active growth of the mycelium, while the short 21 nt RNA would be engaged in the degradation of the target mRNA. Differences in the expression patterns of the two putative Dicer genes and/or different subcelullar locations of their protein products could be responsible for the differential accumulation of the two size classes of antisense RNAs. Alternatively, only 25 nt siRNAs may be produced by the putative Dicer homologue of M.circinelloides, and interaction of the siRNA with the RISC complex would unwind the 25 nt siRNA and would later convert the 25 nt antisense strand into a 21 nt molecule. New experiments are needed to ascertain whether the two classes of antisense RNAs are produced by different Dicer activities and to establish the role of these two classes of RNAs in gene silencing.
Recent results indicate that an amplification step may be required for efficient gene silencing in plants and animals. This step would produce secondary siRNAs using the target mRNA transcripts as a template in a process that would involve a putative RdRP (Sijen et al., 2001; Vaistij et al., 2002). In plants, secondary siRNAs can be detected from sequences upstream and downstream of the initial triggering molecule, indicating bi-directional spreading from the initiator region into adjacent regions of the target gene. Several models have been proposed to explain downstream spreading of RNA targeting, including primer-independent synthesis by a putative RdRP (Tijsterman et al., 2002a). We have demonstrated that endogenous gene silencing in M.circinelloides involves the spreading of RNA targeting, a process that had not been described in fungi. Results indicate that spreading of silencing in M.circinelloides operates at least in the 5′→3′ direction, suggesting a role for a putative RdRP in this organism. Secondary 25 and 21 nt antisense RNAs corresponding to sequences downstream of the input trigger were detected in silenced transformants containing a 5′ portion of the carB gene. Both size classes of antisense RNAs were preferentially produced from the 3′-end of the endogenous gene, which denotes that this so-called ‘transitive effect’ can extend further than 1.5 kb in fungi. Antisense RNAs were hardly detectable with the 5′-end probe, both in the full-length and 3′-truncated carB transformants, indicating that the 25/21 nt molecules detected in silenced transformants correspond mainly to those resulting from amplification on the target mRNA. A preferential primer-independent transcription by the putative RdRP from the 3′-end of the target mRNA and a low processivity of this enzyme could account for the preferential production of small RNAs from the 3′ region of the endogenous gene, as has been proposed in other systems (Zamore, 2002). Recent results indicate that, in plants, the production in vitro of dsRNA by the RdRP activity may be coupled to the production of the 25 nt class of small RNAs (Tang et al., 2003). Our in vivo experiments indicate that, in fungi, both size classes of antisense RNAs are associated with the RdRP-dependent amplification and spreading of silencing signals.
Analysis of the molecular events that differentiate the triggering molecules in silenced and non-silenced transformants, the identification of genes involved in M.circinelloides silencing, and the characterization of the enzymatic activities of the corresponding gene products would allow us to delve more deeply into aspects of the RNA silencing mechanism that are so far unresolved.
Materials and methods
Strains, growth and transformation conditions
The leucine auxotroph R7B (Roncero, 1984), derived from M.circinelloides f. lusitanicus CBS 277.49 (syn. Mucor racemosus ATCC 1216b) (Schipper, 1976), was used as the wild-type strain. Cultures were grown in minimal medium (YNB; Lasker and Borgia, 1980), with or without l-leucine (20 µg/ml), or complete medium (YPG; Bartnicki-Garcia and Nickerson, 1962). The pH was adjusted to 4.5 and 3.2 for mycelial and colonial growth, respectively. Transformation was carried out as described previously (Van Heeswijck and Roncero, 1984) except for the protoplast formation step, which was performed by incubating 2.5 × 108 germinated spores with 15 µg/ml chitosanase RD (US Biologicals) and 0.5 mg/ml Lysing Enzymes (L-1412; Sigma) at 30°C for 45–60 min. Illumination conditions were as described previously (Quiles-Rosillo et al., 2003). Escherichia coli strain DH5α (Hanahan, 1983) was used for all cloning experiments.
Plasmids
Plasmid pMAT647 contains a 2483 bp DNA fragment of the carB genomic sequence, which includes the complete carB gene and regulatory sequences. This fragment was amplified by PCR using the oligonucleotide pairs A (5′-TCGAGAGCTCTGCAGGACAGGACGCCCAGC-3′) and B (5′-GGGGAATTCAGTTAAGGGAGTTAGTGCTAG-3′), which include restriction sites for the enzymes SacI and PstI (primer A, italics) and EcoRI (primer B, italics). The amplified fragment was digested with SacI and EcoRI and cloned into pBluescript II SK+. The resulting plasmid was digested with PstI and ligated to a 4.4 kb PstI fragment of pLEU4 (Roncero et al., 1989). This PstI fragment contains an autonomously replicating sequence in M.circinelloides and the wild-type leuA allele, which complements the leuA mutation present in the R7B strain.
Plasmid pMAT650, which harbours a 677 bp carB genomic fragment that contains the promoter region and 166 bp of the coding sequence, was obtained by ApaI digestion and autoligation of plasmid pMAT647.
Plasmid pMAT754, which contains the complete cDNA sequence of the carB gene under the control of the carB promoter, was kindly provided by J. Gómez-Mateo (University of Murcia).
Plasmid pMAT643 was utilized to prepare the carB RNA probe used in the RNase protection analysis. This plasmid contains a 196 bp fragment of the carB sequence that extends from position +964 to +1159 and includes a 61 bp intron. This fragment was amplified from plasmid pMAT647 by PCR using the primers 5′- AGGAATTCGGACCTGA TTCGCATCAAG-3′ and 5′-CCGAATTCCTGGAGAGATGGCACCT TAGC-3′. Both primers include EcoRI sites (italics) that were used to clone the amplified fragment into the EcoRI site of pBluescript SK+ vector. Restriction analyses and sequencing were performed to identify the correct orientation of the fragment relative to the T7 promoter present in the pBluescript SK+ vector.
Plasmid pMAT645 was used to prepare the pyrG RNA probe utilized as loading control in the RNase protection analysis. This plasmid contains a 158 bp region of the pyrG coding sequence cloned into pBluescript SK+, and was constructed by PCR amplification of a 521 bp fragment from plasmid pEPM9 (Benito et al., 1995) using the primers 5′-CAA ACGTTAACACTTACAAGACTTATAGC-3′ and 5′-CCCTTGATAA TACCTTCTCC-3′. The amplified fragment was digested with HpaI and ApaI enzymes, and the resulting 154 bp fragment was cloned into the vector digested with SmaI and ApaI enzymes.
Plasmids pMAT651 and pMAT652 were used to prepare the carB riboprobes utilized to detect the small carB RNAs in Figures 5 and 6 (Figure 1A, probes a and a′). Both plasmids contain, in different orientations relative to the T7 promoter, a 1662 bp ApaI fragment isolated from plasmid pMAT647 and cloned into the ApaI site of pBluescript SK+. This fragment extends from position +863 to the end of the carB gene. Plasmids pMAT671 and pMAT672 were used to prepare the 5′-end sense and antisense-specific riboprobes (Figure 1A, probes b and b′). They contain a 148 bp NotI fragment isolated from pGCMM20 (Velayos et al., 2000) and cloned, in different orientations, into pBluescript SK+. This fragment extends from position +480 to +628 of the carB gene. Plasmids pMAT675 and pMAT676, used to prepare the 3′-end sense and antisense-specific riboprobes (Figure 1A, probes c and c′), contain a 210 bp PCR-amplified fragment of the carB gene cloned, in different orientations, into pGEM-T (Promega). This fragment extends from position +2224 to +2434 relative to the sequence contained in plasmid pMAT647.
Nucleic acid isolation and analysis
Genomic DNA from M.circinelloides was prepared as described previously (Ruiz-Pérez et al., 1995). Total RNA was isolated using Trizol reagent following the instruction of the supplier (Gibco-BRL). Standard recombinant DNA manipulations were performed as described in Sambrook and Russell (2001) and Ausubel et al. (1989). Southern and northern blots were hybridized to radioactively labelled probes at 65°C in 0.9 M NaCl, 1% SDS, 0.1 g/ml dextran sulfate. Filters (Hybond N+, Amersham Pharmacia Biotech) were washed in 2× SSC, 0.1% sodium dodecyl sulfate (SDS) at room temperature for 5 min, twice in 2× SSC, 0.1% SDS at 65°C for 20 min, and once in 0.1× SSC, 0.1% SDS at 65°C for 20 min.
Probes were labelled with [α-32P]dCTP using Ready-to-Go DNA labelling beads (Amersham Pharmacia Biotech), following the instructions of the supplier. The carB probe used in Figures 1 and 2 corresponds to the 1.8 kb EcoRI–XhoI fragment of the carB cDNA and was isolated from plasmid pGCMM20. The carB probe used in the methylation analysis was a 2.4 kb PstI–XhoI fragment of plasmid pMAT754. This fragment includes the complete carB coding and promoter sequences. The 25S rRNA probe corresponds to a 450 bp fragment containing the 3′ region of the 25S rRNA gene of M.circinelloides. Signal intensities were estimated from autoradiograms using a Shimadzu CS-9000 densitometer.
RNase protection analyses
Total RNA was extracted from mycelia grown in the dark or illuminated with blue light for 4 min at 4 W/m2, and incubated in the dark for 15 min. The antisense RNA probes were prepared by in vitro transcription from BamHI linearized pMAT643 (carB probe) and NotI linearized pMAT645 (pyrG probe), using T7 RNA polymerase. The probes were gel purified before being used in the RNase protection assay. The in vitro transcription reactions and the RNase protection assays were performed using the MAXIscript transcription and RPA III ribonuclease protection assay kits (Ambion) following the instruction of the suppliers. The RNase protection assays were performed using 30 µg of total RNA per reaction.
Isolation and detection of small antisense and sense RNA
Liquid cultures of the wild-type strain and albino transformants grown for different times under continuous illumination conditions were used to isolate small RNAs, following the procedure described by Catalanotto et al. (2002) with minor modifications. Frozen mycelia were pounded in liquid nitrogen and ground to a fine powder in a mortar. Total RNA was isolated using Trizol reagent following manufacturer instructions (Gibco-BRL). The RNA pellets were resuspended in DEPC-treated water and the large RNAs were removed by selective precipitation with 5% polyethylene glycol (PEG) 8000 in 0.5 M NaCl. Small RNAs were precipitated from the supernatants by adding 3 vol. of absolute ethanol and 0.1 vol. of 3 M sodium acetate pH 5, with incubation overnight at –20°C. Small RNAs were resuspended in water and quantified by spectrophotometric analysis.
The small RNAs were then separated by electrophoresis on a 15% polyacrylamide gel (19:1 acrylamide:bisacrylamide) containing 7 M urea in 0.5× TBE buffer, electrotransferred to Hybond N+ filters at 250 mA for 30 min in 1× TBE, and cross-linked by irradiation with ultraviolet (1.2 × 105 µJ/cm2) using a Fluo-Link FLX (Vilber Lourmat). Different oligonucleotides corresponding to sense and antisense sequences of the carB gene were used as size and polarity controls.
Prehybridization was carried out in 40% formamide, 7% SDS, 0.3 M NaCl, 0.05 M Na2HPO4-NaH2PO4 pH 7, 1× Denhardt’s solution and 100 µg/ml sheared and denatured salmon sperm DNA at 30°C for 30 min (Han and Grieson, 2002). Hybridization was carried out in the same solution at 30°C for 16 h. Filters were washed three times in 2× SSC, 0.2% SDS at 50°C for 10 min, and once in 300 mM NaAc, 10 mM Tris–HCl pH 8.0, 5 mM EDTA, 0.81 Kunits/ml RNase A and 20 U/ml RNase T4 at 30°C for 20 min to remove unspecific background. The filters were exposed to Kodak X-OMAT film with an intensifying screen at –70°C.
Sense and antisense-specific riboprobes were prepared by in vitro transcription of linearized plasmids by using the T7 promoter of pBluescript SK+/pGEM-T vectors (MAXIscript transcription kit; Ambion). Riboprobes were treated with RNase-free DNaseI to remove the DNA template and broken to an average size of 50 nt by mixing 20 µl of riboprobe with 300 µl of alkaline buffer (120 mM Na2CO3, 80 mM NaHCO3) and incubating at 60°C for 3 h for the long riboprobes and 2 h for the 5′- and 3′-end probes. Then, 20 µl of 3 M sodium acetate pH 5.0 was added to stop the hydrolysis reaction (Hamilton and Baulcombe, 1999). Low molecular weight RNA marker was produced by RNase T1 digestion of labelled carB antisense transcripts prepared by in vitro transcription of BamHI-digested pMAT643. RNase T1 digestion of the transcript yielded a 35 nt fragment besides several smaller ones.
Acknowledgments
Acknowledgements
We thank Dr V.Garre for his critical reading of the manuscript and helpful discussion, and J.Gómez-Mateo for kindly providing plasmid pMAT754. We also thank J.A.Madrid for technical assistance. This work was funded by the Spanish Dirección General de Investigación, Ministerio de Ciencia y Tecnología (BMC2000-0287). F.E.N. was supported by a graduate fellowship from the Spanish Ministerio de Educación y Cultura.
References
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1989) Current Protocols in Molecular Biology. Green Publishing Associates and Wiley-Interscience, New York, NY. [Google Scholar]
- Bartnicki-Garcia S. and Nickerson,W.J. (1962) Induction of yeast-like development in Mucor by carbon dioxide. J. Bacteriol., 84, 829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito E.P., Campuzano,V., López-Matas,M.A., De Vicente,J.I. and Eslava,A.P. (1995) Isolation, characterization and transformation, by autonomous replication, of Mucor circinelloides OMPdecase-deficient mutants. Mol. Gen. Genet., 248, 126–135. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- Catalanotto C., Azzalin,G., Macino,G. and Cogoni,C. (2002) Involvement of small RNAs and role of the qde genes on the gene silencing pathway in Neurospora crassa. Genes Dev., 16, 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H. (2003) RNA interference: traveling in the cell and gaining functions? Trends Genet., 19, 39–46. [DOI] [PubMed] [Google Scholar]
- Chicas A. and Macino,G. (2001) Characteristics of posttranscriptional gene silencing. EMBO Rep., 2, 992–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C. and Macino,G. (1997) Isolation of quelling-defective (qde) mutants impaired in post-trascriptional transgene-induced gene silencing in Neurospora crassa. Proc. Natl Acad. Sci. USA, 94, 10233–10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C., Ireland,J.T., Schumacher,M., Schmidhauser,T., Selker,E.U. and Macino,G. (1996) Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. EMBO J., 15, 3153–3163. [PMC free article] [PubMed] [Google Scholar]
- Hamilton A.J. and Baulcombe,D.C. (1999) A novel species of small antisense RNA in posttranscriptional gene silencing in plants. Science, 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Hamilton A., Voinnet,O., Chappell,L. and Baulcombe,D. (2002) Two classes of short interfering RNA in RNA silencing. EMBO J., 21, 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S.M., Bernstein,E., Beach,D. and Hannon,G.J. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature, 404, 293–296. [DOI] [PubMed] [Google Scholar]
- Han Y. and Grieson,D. (2002) The influence of inverted repeats on the production of small antisense RNAs involved in gene silencing. Mol. Genet. Genomics, 267, 629–635. [DOI] [PubMed] [Google Scholar]
- Hanahan D. (1983) Studies on transformation of E. coli with plasmids. J. Mol. Biol., 166, 557–580. [DOI] [PubMed] [Google Scholar]
- Ketting R.F. and Plasterk,R.H. (2000) A genetic link between co-suppression and RNA interference in C. elegans. Nature, 404, 296–298. [DOI] [PubMed] [Google Scholar]
- Lasker B.A. and Borgia,P.T. (1980) High frequency heterokaryon formation by Mucor racemosus.J. Bacteriol., 141, 565–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M.A., Mette,M.F. and Matzke,A.J. (2000) Transgene silencing by the host genome defense: Implication for the evolution of epigenetic control mechanisms in plants and vertebrates. Plant Mol. Biol., 43, 401–415. [DOI] [PubMed] [Google Scholar]
- Morel J.B., Mourrain,P., Beclin,C. and Vaucheret,H. (2000) DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr. Biol., 10, 1591–1594. [DOI] [PubMed] [Google Scholar]
- Mourrain P. et al. (2000) Arabidopsis SGS2 and SGS3 genes are required for postranscriptional gene silencing and natural virus resistence. Cell, 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Navarro E., Sandmann,G. and Torres-Martínez,S. (1995) Mutants of the carotenoid biosynthetic pathway of Mucor circinelloides. Exp. Mycol., 19, 186–190. [Google Scholar]
- Navarro E., Ruiz-Pérez,V.L. and Torres-Martínez,S. (2000) Overexpression of the crgA gene abolishes light requeriment for carotenoid biosynthesis in Mucor circinelloides. Eur. J. Biochem., 267, 800–807. [DOI] [PubMed] [Google Scholar]
- Navarro E., Lorca-Pascual,J.M., Quiles-Rosillo,M.D., Nicolás,F.E., Garre,V., Torres-Martínez,S. and Ruiz-Vázquez,R.M. (2001) A negative regulator of light-inducible carotenogenesis in Mucor circinelloides. Mol. Genet. Genomics, 266, 463–470. [DOI] [PubMed] [Google Scholar]
- Pickford A.S., Catalanotto,C., Cogoni,C. and Macino,G. (2002) Quelling in Neurospora crassa. Adv. Genet., 46, 277–303. [DOI] [PubMed] [Google Scholar]
- Quiles-Rosillo M.D., Torres-Martínez,S. and Garre,V. (2003) cigA, a light-inducible gene involved in vegetative growth in Mucor circinelloides is regulated by the carotenogenic repressor crgA. Fungal Genet. Biol., 38, 122–132. [DOI] [PubMed] [Google Scholar]
- Romano N. and Macino,G. (1992) Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol., 6, 3343–3353. [DOI] [PubMed] [Google Scholar]
- Roncero M.I.G. (1984) Enrichment method for the isolation of auxotrophic mutants of Mucor using the polyene antibiotic N-glycosyl-polyfungin. Carlsberg Res. Commun., 49, 685–690. [Google Scholar]
- Roncero M.I.G., Jepsen,L.P., Stroman,P. and van Heeswijck,R. (1989) Characterization of a leuA gene and an ARS element from Mucor circinelloides. Gene, 84, 335–343. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pérez V.L., Murillo,F.J. and Torres-Martínez,S. (1995) PkpA, a novel Phycomyces blakesleeanus serine/threonine protein kinase. Curr. Genet., 28, 309–316. [DOI] [PubMed] [Google Scholar]
- Sambrook J. and Russell,D.W. (2001) Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schipper M.A.A. (1976) On Mucor circinelloides, Mucor racemosus and related species. Studies Mycol., 12, 1–40. [Google Scholar]
- Sijen T. et al. (2001) On the role of RNA amplification in dsRNA-triggered gene silencing. Cell, 107, 465–476. [DOI] [PubMed] [Google Scholar]
- Tang G., Reinhart,B.J., Bartel,D.P. and Zamore,P.D. (2003) A biochemical framework for RNA silencing in plants. Genes Dev., 17, 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M., Ketting,R.F. and Plasterk,R.H.A. (2002a) The genetics of RNA silencing. Annu. Rev. Genet., 36, 489–519. [DOI] [PubMed] [Google Scholar]
- Tijsterman M., Ketting,R.F., Okihara,K.L., Sijen,T. and Plasterk,R.H. (2002b) RNA helicasa MUT-14-dependent gene silencing triggered in C. elegans by short antisense RNAs. Science, 295, 694–697. [DOI] [PubMed] [Google Scholar]
- Vaistij F.E., Jones,L. and Baulcombe,D.C. (2002) Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell, 14, 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heeswijck R. and Roncero,M.I.G. (1984) High frequency transformation of Mucor with recombinant plasmid DNA. Carlsberg Res. Commun., 49, 597–609. [Google Scholar]
- Velayos A., Blasco,J.L., Alvarez,M.I., Iturriaga,E.A. and Eslava,A.P. (2000) Blue-light regulation of phytoene dehydrogenase (carB) gene expression in Mucor circinelloides. Planta, 210, 938–946. [DOI] [PubMed] [Google Scholar]
- Voinnet O. and Baulcombe,D.C. (1997) Systemic signaling in gene silencing. Nature, 389, 553. [DOI] [PubMed] [Google Scholar]
- Zamore P.D. (2002) Ancient pathways programmed by small RNAs. Science, 296, 1265–1269. [DOI] [PubMed] [Google Scholar]
- Zamore P.D., Tuschl,T., Sharp,P.A. and Bartel,D.P. (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell, 101, 25–33. [DOI] [PubMed] [Google Scholar]