Abstract
The three dimensional (3D) structure of the ectodomain of the entire fusion mediating F protein from Sendai virus [MW (trimer) ∼177 kDa] has been determined by cryoelectron microscopy of single molecules and subsequent 3D reconstruction at a resolution of ∼16 Å. The reconstruction, which has been obtained from the native, proteolytic processed fusion primed F1+F2 form, shows the protein protruding ∼170 Å out of the membrane in a homotrimeric association. It consists of a defined ∼65 Å wide distal head and an adjacent neck, which is connected to an 70 Å elongated stalk. Although the overall shape appears to be similar to the recently reported X-ray structure of the Newcastle disease virus F protein, a closer comparison reveals structural differences suggesting that the investigated Sendai F structure represents an advanced state towards the fusion active conformation.
Keywords: 3D reconstruction/cryoelectron microscopy/ F glycoprotein/Sendai virus/single particle analysis
Introduction
The infection of eukaryotic cells by enveloped viruses requires fusion between the viral membrane and the host cell membrane. Specific viral glycoproteins mediate the binding to host cells and/or the fusion with the respective target membrane. For paramyxoviruses, e.g. Sendai virus, Simian parainfluenza virus (SV5), Newcastle disease virus (NDV) or human respiratory syncytial virus (HRSV), two proteins of the envelope bilayer are responsible for the virus entry: (i) the hemagglutinin-neuraminidase protein (HN); and (ii) the homotrimeric fusion protein (F), which is primed to fusion by cleavage of the non-fusion active F0 precursor into the two covalently linked subunits, F1 and F2. HN binds to sialic acids, enabling virus attachment to the host plasma membrane (Scheid et al., 1972; Haywood, 1974), whereas the F1+F2 protein mediates the merger between the viral envelope and the host cell membrane by undergoing a conformational change. For many paramyxoviruses, the F protein mediates membrane fusion only in the presence of their homotypic HN protein. However, neither the trigger mechanism nor the mechanism of the conformational change of the F protein toward a fusion-active state is known.
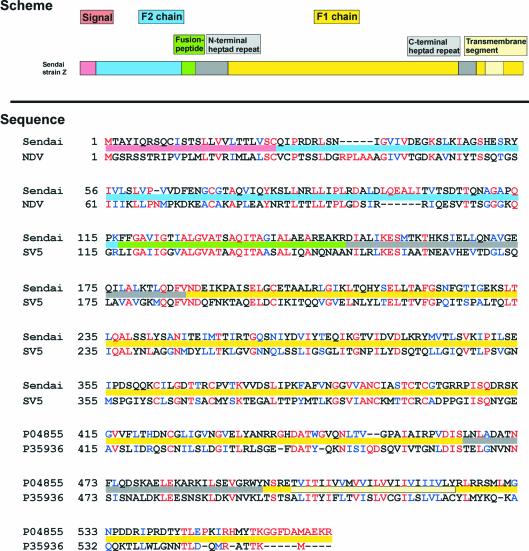
Although the identity of the sequence of F proteins from different paramyxoviruses is limited, segments, which are essential for the fusion process, are highly conserved among Paramyxoviridae (Lamb, 1993) (see Figure 1), such as the hydrophobic N-terminus of the membrane-anchored F1 subunit, the so-called fusion peptide, or the two 4-3 heptad repeat sequences (HR), the C-terminal located HRC and the N-terminal located HRN, which are separated by a relatively long stretch of amino acids (279 residues in the case of Sendai F). It is assumed that these heptad repeat domains refold into a six-fold helix bundle during the fusion process, as supported by the crystal structure of fragments of the F1 subunit of HRSV F (Zhao et al., 2000) and SV5 F (Baker et al., 1999). The HRN segment forms a trimeric coiled-coil, which is surrounded by three antiparallel helices of the HRC. Consequently the transmembrane domain and the fusion peptide, which is known to insert into the target membrane (Scheid and Choppin, 1974, 1977; Gething et al., 1978; Novick and Hoekstra, 1988), are both transposed into close association. It has been suggested that this structural state corresponds to a fusion-competent conformation, bringing the viral envelope and the target membrane into close apposition (Ben-Efraim et al., 1999; Baker et al., 1999; Melikyan et al., 2000; Peisajovich et al., 2000; Zhao et al., 2000; Russell et al., 2001). Indeed, soluble derivatives of HRN and HRC inhibit fusion, presumably by binding to the HRC or HRN domain of the F protein, respectively (Rapaport et al., 1995; Joshi et al., 1998; Ghosh and Shai, 1999; reviewed in Lamb and Kolakofsky, 2001; Russell et al., 2001). However, it remains open whether the formation of the six-fold helix bundle is a prerequisite, a stimulus, or the result of the process.
Fig. 1. Primary structure of the F protein of the paramyxoviruses Sendai virus and NDV. Activation of the precursor F0 is initiated by enzymatic cleavage into two subunits, F1 and F2. Preserved sequences of the F1 subunit are the fusion sequence and the two heptad repeat domains, HRN and HRC. The coloured bars within the sequence text identify the regions as defined by the scheme above (corresponding to Sendai F).
Recently, the three-dimensional (3D) structure of the recombinantly expressed ectodomain of the trimeric F protein from NDV has been determined by X-ray crystallography at a resolution of 3.3 Å (Chen et al., 2001a). The ectodomain can be differentiated into a distal head, a neck and a stalk region proximal to the viral membrane. The head, which consists of two β-domains (the ‘β barrel domain’ and the ‘immunoglobulin-like β domain’), and the upper part of the neck are both penetrated by an axial channel. In the plane of the head–neck interface this axial channel trifurcates into three radial channels, thus creating a distinct central cavity. The most striking feature of the neck and the stalk is a central triple-helix coiled-coil, composed of residues G171-F221. Residues <171 (including the fusion peptide) and >454 (including the HRC) could not, however, be localized. This has been attributed to proteolysis of the initially used F0 protein upon crystallization, suggesting that the presented structure may correspond to a mixture of F0 and a cleaved form of the F protein (Chen et al., 2001a,b).
For fusion-primed proteins, the 3D structure of the complete ectodomain is known only in a few cases: hemagglutinin (HA) of influenza virus A (Wilson et al., 1981), hemagglutinin–esterase fusion glycoprotein (HEF) of influenza virus C (Rosenthal et al., 1998), envelope glycoprotein E of the tick-borne encephalitis virus (TBEV) (Rey et al., 1995), and envelope glycoprotein E1 of Semliki Forest Virus (SFV) (Lescar et al., 2001). To understand the mechanism of viral membrane fusion, however, the conformational pathway of the entire fusion mediating protein, from the non-fusion activated state via the fusion-primed to the fusion active state, must be understood.
Cryoelectron microscopy in combination with single particle 3D-structure determination allows identification of protein conformations without the need to generate crystals for X-ray analysis. A multitude of 3D macromolecular structures have now been routinely elucidated using this method (e.g. Baumeister and Steven, 2000; Mancini et al., 2000; Zhou et al., 2001). Previously, we have shown that the method is even suitable for characterization of protein structures in different conformational states. The 3D structure of the influenza virus A hemagglutinin ectodomain has been elucidated for a fusion-primed, as well as for a fusion-competent conformation at a resolution of 10 and 14 Å, respectively (Böttcher et al., 1999).
In the present study we report the 3D structure of the intact ectodomain of the Sendai F protein in its cleaved, fusion-primed F1+F2 state at a resolution of 16 Å using cryoelectron microscopy and 3D reconstruction techniques of single particles. With a molecular weight of 176.7 kDa, the Sendai F-protein is so far one of the smallest protein structures solved by this method. The protein appears to be of a ‘mace’-like shape, protruding ∼170 Å out of the membrane. At first sight it resembles typical overall features of the NDV F protein and descriptively consists of a defined ∼65 Å wide distal head and an adjacent neck, which is connected to a 70 Å elongated stalk. A closer comparison, which will be discussed in detail, however, indicates that the investigated Sendai F protein might represent an advanced state towards the final fusion conformation.
Results
Characterization of purified F1+F2 protein
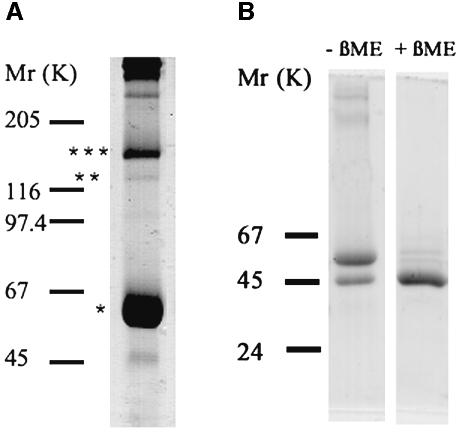
1The protein was purified from detergent extracts of Sendai viruses (strain Z). SDS–PAGE followed by Coomassie Brilliant Blue staining revealed specific isolation and enrichment of the F protein (Figure 2). Based on the sequence of the F protein, the molecular weights of the F1+F2 monomer, dimer and trimer are 58.9, 117.8 and 176.7 kDa, respectively, and that of the F1 monomer is 49 kDa. A major band at ∼60 kDa and a band of low intensity at ∼48 kDa corresponding to monomeric F1+F2 and F1 (Figure 2A, lane 1), respectively, were found under non-reducing conditions. Furthermore, at 1% SDS (Figure 2A, lane 1) we detected a faint band at ∼120 kDa and a more intense band at 160 kDa, corresponding to the dimer and trimer of F1+F2, respectively, but not at 2% SDS (data not shown). Obviously, a SDS concentration of 1% was not sufficient to disrupt non-covalent oligomerization of the F1+F2 protein completely. Under reducing conditions, the disulfide bridges between F1 and F2 were broken, the major band at 60 kDa disappeared, and the prominent F1 band was detected (Figure 2B). Because of its rather low molecular weight of 9.9 kDa, the F2 subunit passed through the gel and could therefore not be detected. Notably, in no case we did find any indication for the presence of the precursor F0 in our preparations. Thus, the 3D structures found for the ectodomain (see below) correspond to the F1+F2 protein.
Fig. 2. Analysis of purified F protein from Sendai virus. The F protein was isolated and reconstituted as described in Materials and methods. The probes were analysed by SDS–PAGE under non-reducing conditions [(A) 7.5% gel (1% SDS)] and reducing conditions [(B) 10% gel (3% SDS)] and stained with Coomassie Brilliant Blue. Under non- reducing conditions a major band (*) was observed at ∼60 kDa, corresponding to the F1+F2 protein (A). Bands of dimers and trimers of (F1+F2) are indicated by ** and ***, respectively. Under reducing conditions [(B) right lane (+βME, with β-mercaptoethanol], the band of F1 at 50 kDa becomes visible, while the F1+F2 band seen in the left lane (–βME, without βME) disappeared. Molecular mass markers, given in kiloDaltons, are shown on the left. The molecular weight of the visualized protein bands was determined using Software Sigma-Scan Image.
Fusion and hemolytic activity of virosomes
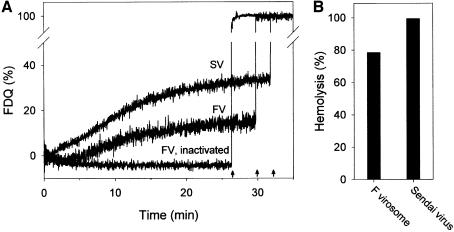
To determine whether the isolation procedure affects the function of the F protein, we measured its hemolytic and fusion activity upon reconstitution into liposomes and compared them with those of virions (Figure 3). For fusion, we used HepG2 cells because it has been shown previously that F protein from Sendai virus mediates fusion with the plasma membrane of those cells, even in the absence of the HN protein (Bagai et al., 1993). It has been suggested that HepG2 cells bear a surface glycoprotein receptor (asialoglycoprotein), which enables the F protein to bind and subsequently to mediate fusion with the plasma membrane of HepG2 (Bagai et al., 1993). Virosomes and Sendai virions contained the fluorescent phospholipid analogues Rh-PE and R18, respectively, at self-quenching concentrations. After binding of virosomes to HepG2 cells at 4°C, fusion was initiated by incubation of virosome–cell complexes at 37°C as indicated by an increase in fluorescence (Figure 3A). Upon fusion, redistribution of Rh-PE to the plasma membranes of HepG2 cells is accompanied by a relief of self-quenching. Thus, the increase in fluorescence intensity is a direct measure of fusion. The fusion of virosomes with HepG2 cells, as well as the hemolytic activity of virosomes were comparable to those of intact Sendai virions with HepG2 cells (Figure 3A and B), indicating that neither activity of the F protein was affected by isolation and reconstitution procedures. However, fusion of Sendai virus was faster and the extent to which it occurred slightly higher. Presumably, the presence of the HN protein promoted the fusion of Sendai virions with HepG2 cells with respect to F virosomes lacking HN (Bagai et al., 1993). Fluorescence dequenching is not due to a non-specific transfer of fluorescent analogues between virosomes (virions) and HepG2 cells, because no dequenching was observed when virosomes were inactivated for 20 min at 56°C (Figure 3A, only shown for F virosomes). It is known that the fusion activity of the F protein of Sendai virus is inactivated by incubation at temperatures >50°C (Wharton et al., 2000). Similarly, the hemolytic activity of F virosomes and Sendai virions was abolished after heat treatment (not shown).
Fig. 3. (A) Fusion kinetics of reconstituted F protein virosomes (FV) or intact Sendai virus (SV) with HepG2 cells at 37°C, pH 7.4. Rh-PE-labelled virosomes or R18-labelled Sendai virions were bound to HepG2 cells for 30 min at 4°C. Subsequently, fusion was initiated (t = 0 min) by transfer of virosome cell complexes to pre-warmed buffer (37°C). Fusion is measured by the relief of self-quenching of Rh-PE and R18 of FV and SV, respectively, caused by redistribution of the fluorescent lipid analogue to the plasma membrane of HepG2 cells. Fusion was completely inhibited when F virosomes (FV, inactivated) or Sendai virions (not shown) were incubated for 20 min at 56°C. Fluorescence dequenching (FDQ) is normalized to the fluorescence intensity obtained after addition of 0.5% Triton X-100 at the end of the kinetics (see arrows, infinite dilution of the analogue). (B) Haemolytic activity of F virosomes and Sendai virions after incubation with red blood cells for 30 min at 37°C.
Electron microscopy and image analysis
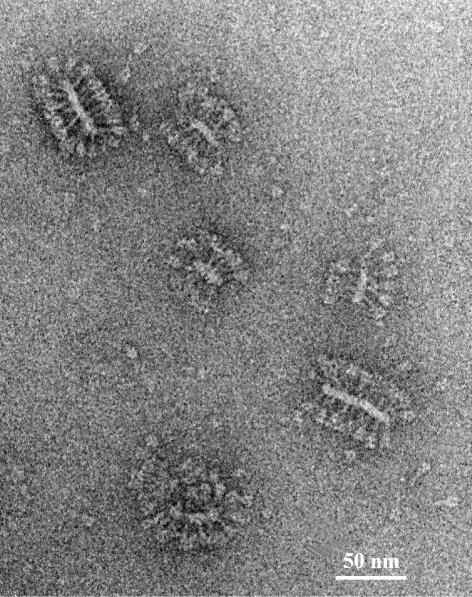
An electron micrograph representing the typical appearance of the reconstituted F protein is shown in Figure 4. Upon initial visual inspection of the electron micrographs, Sendai F protein molecules appear as ‘mace’-like spikes protruding from the filamentous lipid vesicles. The overall appearance of such assemblies resembles that of native virosomes.
Fig. 4. Electron micrograph of a cryo-negative stain preparation [containing phospho-tungstic acid, 1% (w/v)] of purified and reconstituted F protein. After reconstitution, Sendai F protein trimers are predominantly anchored in filamentous lipid vesicles, i.e. the membrane appears as two elongated, closely packed bilayers in the projection images. Occasionally, non-anchored single trimers occur (e.g. bottom right). Bar = 50 nm.
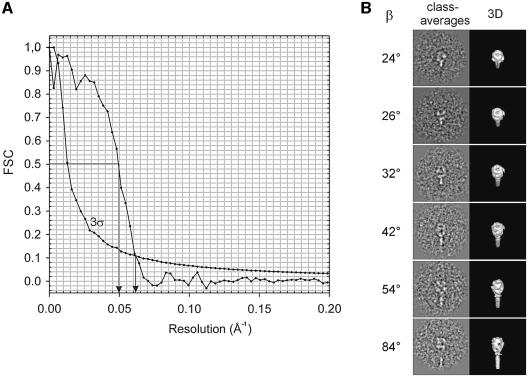
Based on a data set of 3500 extracted single molecular images, the 3D structure of the Sendai F protein was reconstructed at a resolution of ∼16 Å (3σ threshold, see Figure 5A). The preliminary steps were done without imposing symmetry. After several iterative procedure cycles it became evident that three-fold symmetry would match the structure. A selection of representative class averages varying in ‘tilt angle’ (beta) is presented in Figure 5B.
Fig. 5. (A) Fourier Shell Correlation (FSC) of the final Sendai F protein 3D reconstruction, plotted as function of spatial frequency, together with the 3σ threshold curve (corrected for the C3 pointgroup symmetry). Resolution values corresponding to the 0.5 criterion and 3σ threshold, respectively, are marked by arrows. (B) Representative class averages together with the corresponding 3D surface representations at different ‘tilt’ angles β.
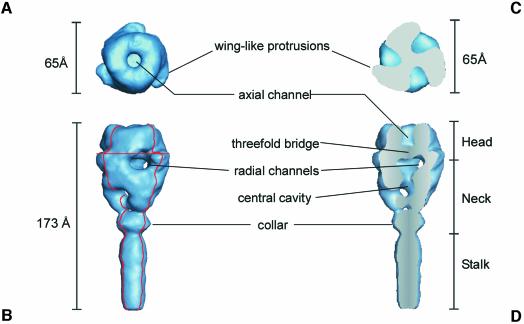
The final 3D reconstruction of the F ectodomain is shown in Figure 6. The total length of the ‘mace-like’ ectodomain of F1+F2 is 173 Å. The distal ‘head’ of the ectodomain is ∼65 Å in both length and width. The top view of the head reveals three wing-like protrusions. The head harbours axial and radial channels. The axial channel is separated from the radial channels by a massive three-fold bridge, which is located ∼27 Å from the top of the head. The radial channels are connected to a central, low-density domain of the neck region (central cavity). The head is connected by a distinct ‘neck’ region to a 70 Å ‘stalk’ of the ectodomain. Proximal to the stalk, the neck region forms a collar.
Fig. 6. Surface representations of the 3D reconstruction of the ectodomain of Sendai virus F protein determined from cryoelectron micrographs at a resolution of ∼16 Å. (A) Top view; (B) side view. The viral/virosome membrane (not shown) is located at the lower end of the the stalk region. (C) Top view of a horizontal section [the localization of the cross section is indicated by the horizontal red line in (B)]. (D) Axial cut-away view (45°) of (B) [red contours in (B) indicate the localization of the cutting planes]. Various structural details of the F ectodomain are indicated (for further description see Results and Discussion).
Docking of Sendai F and NDV F ectodomain structure
In order to enable direct comparison of the Sendai F and NDV F ectodomain structures, we applied a Gaussian filter to the crystal data of NDV F to remove high-resolution information. Both structures are shown at a resolution level of 16 Å in Figures 6 and 7A, respectively.
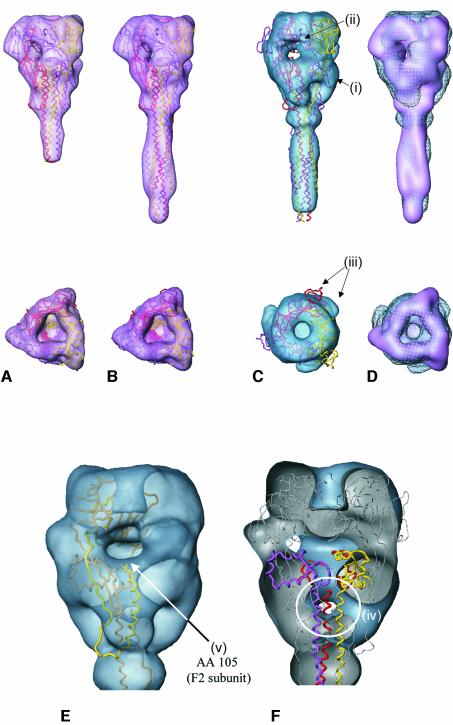
Fig. 7. (A–D) Side views (top row) and top views (bottom row) of the NDV F protein X-ray structure (A) and of a chimeric construct composed of the X-ray structure of the NDV F protein (Chen et al., 2001a), and the X-ray structure of a fragment taken from the SV5 F (six-helix-bundle motif) (Baker et al., 1999) (B–D), respectively, in comparison with the 3D reconstruction of Sendai F protein (for details see Discussion). (A) The X-ray structure of NDV F at high resolution (backbone representation; monomers coloured magenta, yellow and red) and gaussian filtered at 16 Å resolution (surface presentation, magenta), respectively. (B) X-ray structure of the chimeric NDV/SV5 F structure at high resolution (backbone presentation; monomers in magenta, yellow and red) and gaussian filtered at 16 Å resolution (surface representation, magenta), respectively. (C) Three-dimensional structure of the chimeric NDV/SV5 F structure (backbone representation) docked into the Sendai F density map using Situs software. The Sendai F structure (surface presentation, blue) as obtained from cryoelectron micrographs at 16 Å resolution. For structural details (i–iii) see Discussion. (D) Surface representations of the docked low pass filtered NDV/SV5 F structure (shaded surface) in comparison with the Sendai F EM structure (meshed surface). (E–F) Fitting of NDV and Sendai F reveals distinct differences between both structures: (E) One monomer of the chimeric NDV/SV5 F structure (backbone presentation) docked into the Sendai F density map (head/neck region shown in transparent surface representation) using Situs software. According to this docking, the onward extension of F2 starting at amino acid 105 (arrow), which is missing in the crystal structure of NDV F and is present in the Sendai F structure, would be localized in a low-density region of the Sendai F protein. (F) Chimeric NDV/SV5 F structure (backbone presentation) docked into the Sendai F density map (head/neck region shown as cut surface presentation) using Situs software. The domain of relatively high density in the central part of the NDV neck, which correlates to tightly associated central helices (amino acids 220 to 171), does not fit well with the low-density area of Sendai F protein in this region (circled).
A comparison of the isosurfaces of both structures is not sufficient to achieve a quantitative insight into the extent to which the two structures coincide in three dimensions. We therefore employed the Situs software package, which performs a docking procedure of two structures on the basis of 3D density maps (Wriggers and Birmanns, 2001). However, docking of NDV F (Figure 7A) into the Sendai F structure (Figure 6) using the Situs-program CoLoRes resulted at first glance in a sound fit, but revealed several deviations in detail (not shown). One of the major doubts we had with this result was based on the fact that the docking might have been affected by the missing stalk of the NDV F structure. We therefore found it advantageous to compare the structures on the basis of a similar overall structure. To address this concern, we followed a superposition model suggested by Chen et al. (2001a), where the missing part of the stalk of NDV F protein has been complemented by extending the crystal structure with the six-helix bundle motif from the SV5 F protein (Baker et al., 1999).
The chimeric NDV/SV5 F structure (Figure 7B) has been constructed and kindly provided by Dr Brian Smith (CSIRO Health Sciences and Nutrition, Parkville, Australia). The superposition of SV5 F and NDV F is based on a segment of 28 common residues in the HRN section of both structures. The corresponding antiparallel HRC segment (including the extension up to the transmembrane domain) is missing in the NDV F structure, while it is present in the SV5 six-helix bundle. Although there is no evidence for the existence of a six-helix bundle motif in the crystallized structure of NDV F, this chimeric construct should allow near modelling of the length of the missing part of the NDV F protein stalk, and therefore the full length of the NDV F ectodomain. This composite structure (see Figure 7B, gaussian filtered to a resolution comparable to electron microscopy) was taken for a second docking experiment using Situs ‘qrange’. The result is shown in Figure 7C and D.
Discussion
The 3D reconstruction of the Sendai F glycoprotein, which has been determined in its cleaved, fusion-primed F1+F2 form, revealed a homotrimeric organization of the F protein. This finding is in agreement with previous studies showing the trimeric organization of the F protein in other paramyxoviruses such as SV5 (Russell et al., 1994), HRSV (Calder et al., 2000) and NDV (Chen et al., 2001a). The overall appearance of the Sendai F ectodomain is similar to that of HRSV F obtained using electron microscopy (Calder et al., 2000; see below) and to the recently reported X-ray structure of NDV F (Chen et al., 2001a).
Comparison of the Sendai F and NDV F ectodomain structure
A comparison of the primary structure of both proteins, Sendai F and NDV F, reveals a sequence identity of ∼25% and a sequence similarity of ∼40%, both of which are relatively high values for viral fusion proteins. For example, in the case of the very similarly folded fusion-mediating E1 (SFV) (Lescar et al. 2001) and E proteins (TBEV) (Rey et al. 1995), sequence identity is only 12%. A sequence comparison between the Sendai F protein and NDV F also reveals that the identities are homogeneously distributed over the entire protein, so that similar 3D structures are expected for both F proteins. Aside from obvious differences in the length of the stalk, the isosurface-rendered structures of NDV F and Sendai F reveal remarkably good agreement in spatial organization at this level of resolution (Figure 6 and 7A). This is evident from the overall shape and dimension of both head domains. As is obvious from the top view, both structures are characterized by a triangular arrangement and a central opening. The head domain of Sendai F harbours a noticeable pattern of radial and axial channels, similar in size and position to that of NDV F.
To obtain a detailed and quantitative comparison, a docking procedure of both structures was undertaken (Figure 7). To overcome the problem of fitting two structures with entirely different centres of mass, a chimeric structure was deployed, complementing the missing part of the stalk in the NDV F structure (Chen et al., 2001a) by extending the NDV F crystal structure using the six-helix bundle motif of the SV5 F protein (see Results). At first glance, features such as the overall dimensions, the location of the axial channel and the length of the stalk appear to be in good agreement. However, closer examination of the fitted structures reveals several differences: (i) Sendai F protein shows additional density in the outer parts of the neck (Figure 7C, i); (ii) the connection between the radial and axial channels observed for NDV is interrupted by a massive central bridge in the case of Sendai F (Figure 7C, ii); (iii) the three outer wing-like protrusions of Sendai F are somewhat twisted against NDV F (Figure 7C, iii); (iv) the domain of relatively high density in the central part of the NDV neck corresponding to the tightly associated central helices (aa 220 to aa 171) does not fit with the low density area of Sendai F protein in this region (Figure 7F, circle); and (v) the onward extension of F2 starting at amino acid 105 that is missing in the crystal structure of NDV F would be precisely localized in a low-density region of the Sendai F protein (Figure 7E, arrow).
The structures of Sendai F and NDV F reflect a different conformation of the F ectodomain
To summarize, although the docking experiment shows that NDV F and Sendai F adopt a similar overall spatial organization, distinct differences exist between both 3D structures. We therefore surmise that both proteins represent different conformational states along the fusion pathway. This argument is strengthened by considering recent low-resolution studies on the morphology of the F protein. Calder et al. (2000) reported two different conformations of the HRSV F ectodomain derived from the raw images of negatively stained and dried samples: a cone-shaped and a lollipop-shaped state. Subsequent studies revealed that the non-cleaved precursor F0 adopts the cone-shaped morphology, whereas the formation of the lollipop conformation requires an enzymatic cleavage of F0 (González-Reyes et al., 2001; Ruiz-Argüello et al., 2002).
One may wonder how the 3D structure of Sendai F groups into this morphological classification, and how this compares with NDV F. The electron microscopy-based morphological judgement by Calder et al. (2000) resulted from simple visual inspection of individual projections of single randomly orientated particles. However, alignment and classification routines of images should allow this visual differentiation to be objectified. To enable a direct comparison, we calculated side-view projection images (β = 90°) of filtered NDV F X-ray data and of the Sendai F1+F2 electron density map (not shown). Indeed, morphologically the NDV F corresponds to a cone-like shape, as suggested by Chen et al. (2001a), whereas the Sendai F protein adopts a more lollipop-like shape. This classification is in agreement with that of Calder et al. (2000), assigning the precursor F0 to the cone-like shape and the cleaved form F1+F2 to the lollipop-like shape (see above).
Cleavage of F0 triggers a conformational change: implications for F protein-mediated fusion
Based on the observation that the cleavage of the paramyxovirus F protein is accompanied by a morphological change from a cone- to a lollipop-shaped structure, and that the NDV F structure corresponds to a cone whereas the Sendai F1+F2 adopts the lollipop form, we conclude that the cleavage of Sendai F0 is accompanied by structural alterations to the ectodomain. Due to the high identity of the primary structure, we consider it unlikely that structural differences are based on sequence differences.
There are several lines of evidence showing that upon cleavage of the F0 protein to F1+F2, conformational changes occur, as has been shown for Sendai virus (Hsu et al., 1981), NDV (Kohama et al., 1981; Umino et al., 1990) and SV5 (Dutch et al., 2001). Circular dichroism spectroscopy showed alterations in the conformation between the precursor and the cleaved F protein of Sendai virus, and an exposure of hydro phobic domains was also found (Hsu et al., 1981). Significant changes in F protein antibody recognition for SV5 were detected after cleavage of the precursor F0 protein to the fusion-active disulfide-linked heterodimer F1+F2 (Dutch et al., 2001). Antibodies directed against the heptad repeat regions recognized those regions only in the uncleaved form.
Cleavage of HRSV F at two distinct sites is required for the activation of membrane fusion (González-Reyes et al., 2001). While one site is highly conserved among paramyxoviruses, the second site is found only for HRSV F. Fusion activation is accompanied by changes in the HRSV F structure from cone- to lollipop-shaped spikes, as shown by electron microscopy (González-Reyes et al., 2001). However, this conformational change is observed only when the highly conserved site is cleaved (Ruiz-Argüello et al., 2002). This implies that the acquisition of membrane fusion activity triggered by the cleavage and the change of the F protein shape are directly linked (González-Reyes et al., 2001). It has been suggested that the cone-shaped conformation may correspond to a pre-fusion state, while the lollipop-shaped form might represent a later fusion-activated state or even post-fusion state (Calder et al., 2000). Chen et al. (2001a) proposed that the lollipop-shaped conformation might in general correspond to a post-fusion state. However, considering the parallel assessment of fusion activity, we can clearly state that the Sendai F1+F2 protein preparation must be assigned to a fusion-primed but not to a post-fusion state. The latter observation does not necessarily preclude that the post-fusion state of the F protein corresponds to a lollipop-like structure. It rather suggests that different intermediates of the cleaved F protein can adopt such a morphology, and that the characterization of the overall shape of the ectodomain is not sufficient to distinguish between the pre- and the post-fusion state.
Once cleavage has occurred, the F protein is primed for fusion and undergoes further conformational changes towards a pre-hairpin intermediate upon interaction with the host cell receptor or, if necessary, with the HN protein (Russell et al., 2001). In this state the N-terminus of F1, including HRN, is released from the head domain towards the target membrane, and the fusion sequence eventually inserts into the target membrane. The movement of the fusion sequence toward the target membrane may be supported by the formation of a coiled-coil arrangement of the HRN sequences (Singh et al., 1999). In a next step, the pre-hairpin intermediate transforms into a hairpin structure characterized by the binding of the HRC to the HRN coiled-coil, forming a six-helix bundle in close proximity to the viral membrane. This conformational alteration results in membrane apposition, enabling the final fusion of both membranes. It has been suggested that the free energy associated with the conformational rearrangements of the F protein could be coupled directly to the membrane fusion event (Baker et al., 1999; see also Melikyan et al., 2000). Thus, formation of the six-helix bundle is essential for late steps of fusion.
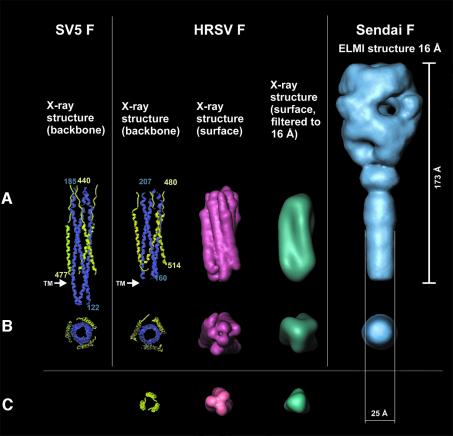
The Sendai F protein used in this study is capable of mediating fusion, implying that the helix bundle has not been formed at this stage. However, based on NMR studies of the 3D solution structure of the ectodomain of gp41 of SIV, Caffrey et al. (1998) suggested that the heptad repeat containing viral proteins, which trigger fusion at neutral pH, may exist in an equilibrium between a non-associated form and the six-helix bundle form. Thus, one wonders whether the reconstructed ectodomain of Sendai F1+F2 protein may already comprise the six-helix bundle. We have therefore compared the size of the stalk of Sendai F protein with the six-helix bundle of SV5 (Figures 7 and 8) and HRSV (Figure 8) at a resolution of 16 Å. In Figure 8, the backbones of the X-ray structures of the six-helix bundle of SV5 and HRSV are aligned to the cryoelectron microscopy structure, so that the respective transmembrane domains are in the comparable position (see arrows in Figure 8). Although the diameter of the six-helix bundle seems to be somewhat larger, we cannot provide an unambiguous answer regarding whether the cleaved and fusion-primed Sendai F protein harbours a six-helix bundle or not. Unless the packing parameters of the HRC helices are known, a simple comparison of the stalk diameter is not sufficient and further studies on atomic resolution are necessary. Nevertheless, it has been demonstrated that the formation of the core complex is essentially a non-reversible process. Even after treatment at 100°C, the six-helix bundle is still resistant to proteases (Dutch et al., 2001). We therefore consider it unlikely that the six-helix bundle forms the stalk of the F1+F2 ectodomain reconstructed in this study.
Fig. 8. Crystal structures of fragments of the SV5 F and of the HRSV F, respectively, in comparison with the 3D structure of the Sendai F ectodomain determined by cryoelectron microscopy (see Figure 5). From left to right (all structures at the same scale): (A) backbone representation of the X-ray structure of a SV5 F fragment (Baker et al., 1999) (N-helices are shown in blue, C-helices in yellow); backbone representation of the X-ray structure of a HRSV F fragment (Zhao et al., 2000); surface representations of this structure at high resolution (light red) and filtered at 16 Å resolution (turquoise); and 3D structure of the Sendai F ectodomain (16 Å resolution) determined from cryoelectron micrographs (blue). (B) Bottom view of the structures presented in (A). The head domain of the 3D Sendai F structure is omitted for clarity. (C) A three-fold helix in backbone (yellow), surface (light red) and low-pass filtered (16 Å resolution) surface representation (turquoise).
In conclusion, the very similar structure of the F protein of NDV and Sendai virus suggests a common spatial organization of paramyxovirus F proteins. Priming the F protein for fusion by cleavage of its precursor F0 is accompanied by a significant conformational change, which appears morphologically as a transition from a cone-like towards a more lollipop-shaped ectodomain. The corresponding structural refolding can be described as an ‘iris’-like mechanism, where the outer ‘wings’ allow space for the opening of a central cavity. Such a mechanism has been proposed previously for influenza HA when adopting a fusion-competent ectodomain structure (Böttcher et al., 1999). The massive central bridge in the head of the cleaved Sendai F corresponds to the predicted position of the fusion peptide in the pre-fusion state of NDV F (Chen et al., 2001a). Thus, to expose the fusion peptide, the fusion-primed ectodomain has to undergo further conformational changes.
Materials and methods
Isolation, purification and reconstitution of F protein
Sendai virus (strain Z) was grown for 48 h in 10-day-old embryonated hen eggs. The F protein was purified according to Tomasi and Loyter (1981) with minor modifications, described by Bagai et al. (1993). The purity of F virosomes was checked by SDS–PAGE, with the SDS gel under reducing and non-reducing conditions. Gels were stained with Coomassie Brilliant Blue. The molecular weight of the visualized protein bands was determined using Software Sigma-Scan Image.
For the reconstitution of F protein, phosphatidylcholine from egg yolk (eggPC; Sigma) and phosphatidylethanolamine-N-(lissamine rhodamine B sulfonyl) (Rh-PE; Avanti Polar Lipids, Inc.) were used. PC (130 nmol) and 56 nmol Rh-PE dissolved in chloroform were dried in a glass vial under nitrogen to form a thin film. The supernatant from the detergent extract, containing only the viral proteins and lipids, was added to the lipid film and incubated at RT for 15 min with gentle shaking. The detergent was removed by SM2 Bio-Beads (Bio-Rad Laboratories) according to the manufacturer’s instructions.
Cell culture
Monolayer cultures of HepG2 cells (American Type Culture Collection, Rockville, MD) were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% (vol/vol) fetal calf serum, 2 mmol/l glutamine and antibiotics (100 IU of penicillin and 100 mg/l of streptomycin) (Biochrome KG) as described by Müller et al. (1996). For fusion measurements, cells were resuspended in Dulbecco’s phosphate-buffered saline (PBS) containing 2 mM Ca2+ and 2 mM Mg2+ (DPBS+).
Labelling of intact Sendai virus
Sendai virus was labelled with the fluorophore octadecylrhodamine B chloride R18 (Molecular Probes) as described previously (Hoekstra et al., 1984).
Fusion assay
Fusion of Rh-PE-labelled virosomes or R18-labelled Sendai virions with HepG2 cells was measured at pH 7.4 and at 37°C as described previously (Bagai et al., 1993). To inhibit endocytotic uptake of virosomes, buffer contained 20 mM NaN3 to inhibit endocytosis. After 45 min, 0.5% Triton X-100 was added to obtain maximal dequenching (Fmax). Fusion was monitored and quantified by fluorescence dequenching (FDQ) (Blumenthal et al., 1987) using an AMINCO BOWMAN 2 fluorescence spectrometer at 560 and 590 nm excitation and emission wavelengths, respectively.
Hemolysis
Human erythrocytes (RBCs) (DRK) were washed three times with PBS and resuspended to a 2.5% haematocrit. After 500 µl of RBCs were mixed with 50 µl of F virosomes, wheat germ agglutinin (WGA) was added to a final concentration of 8 µg/ml. The suspension was incubated on ice for 30 min. After 30 min at 37°C with periodic shaking, 50 µl of the virosome–erythrocyte suspension was transferred into 1 ml of chilled PBS to terminate the reaction, and spun at 13 000 g for 5 min. The amount of haemoglobin released in the presence of 1 mM Triton X-100 was taken as 100% haemolysis. As control for spontaneous and WGA-induced leakage, virosomes were pre-treated at 56°C for 20 min. Sendai virus-induced haemolysis was obtained as described above, but without the addition of WGA.
Electron microscopy
Droplets of the sample (5 µl) were applied to hydrophilized carbon-covered microscopical coppergrids (400 mesh) and supernatant fluid was removed with a filter paper until an ultrathin layer of the sample was obtained. Again, a droplet of contrasting material (1% phosphotungstic acid at pH 7.4) was added and blotted. A heavy metal-stained layer of the protein assemblies was thus prepared, which has been subsequently plunged into liquid ethane in order to preserve the protein structure in its hydrated state.
The Gatan-626 specimen holder/cryo-transfer system was used in a Tecnai F20 FEG transmission electron microscope (TEM; FEI Company). The sample temperature was maintained at –180°C. Imaging was performed under low-dose conditions using a primary magnification of 51064× at an accelerating voltage of 160 kV. The defocus value was chosen to correspond to a first zero of the CTF at ∼9 Å.
This specimen preparation procedure was essentially the same described previously for the 3D structure determination of influenza HA (Böttcher et al., 1999). An embedding matrix with a somewhat higher contrast compared with the conventional vitreous-ice preparation is obtained. Despite the relatively high acceleration voltage of the microscope and the chosen ‘close-to-focus’ imaging conditions, the signal-to-noise ratio is improved and the beam sensitivity is reduced (de Carlo et al., 2002). Using this preparation technique, it has been shown by comparison of both the X-ray crystal structure and the EM reconstruction of HA that there is no significant structural difference within the limits of the achieved resolution of 10 Å.
Reconstruction of the 3D structure
Laser-optically checked micrographs were digitized using the Heidelberg Primescan drum scanner (Heidelberger Druckmaschinen AG) at a nominal pixel resolution of 0.78 Å in the digitized images.
All image processing was performed using the Imagic-5 software package (IMAGE Science GmbH). Molecules were interactively selected and extracted from the digitized micrographs as 400 × 400 pixel fields. For computational efficiency these single images were interpolated to 1.56 Å pixel size for all subsequent steps. The 3D reconstruction was calculated using the ‘angular reconstitution’ approach (van Heel, 1987).
Fourier shell correlation (FSC) (van Heel and Harauz, 1986) of two different 3D reconstructions, each of which included half of the final class averages, was done to assess the resolution. For the 3D structure the resolution obtained in the final 3D reconstruction was determined to be ∼16 Å applying the 3σ threshold (∼21 Å at 0.5 criterion), taking C3 point-group symmetry into account (Orlova et al., 1997).
Fitting of atomic structures into EM density maps
The Situs Software Package Version 2.0 (http://situs.scripps.edu/index.html) was used to fit the high-resolution X-ray structures into the EM density maps. The docking procedure was performed as described by Wriggers and Birmanns (2001).
Acknowledgments
Acknowledgements
We are indebted to Dr Holger Stark (Max-Plank-Institut für Biophysikalische Chemie, Göttingen, Germany) for stimulating discussions, and Bärbel Hillebrecht for virus growing. In particular, we are indebted to Dr Mike Lawrence (CSIRO Health Sciences and Nutrition) for critical discussions and his suggestion of using the ‘chimeric construct’. We also thank Dr Brian Smith (CSIRO Health Sciences and Nutrition) for providing us with chimeric data. R.Schmidt and Dr M.Schatz (IMAGE Science GmbH, Berlin, Germany) are gratefully acknowledged for their continuous technical support. We are grateful to the Deutsche Forschungsgemeinschaft for the Philips Tecnai F20 TEM and generous support. B.B. is a recipient of the Heinrich-Böll-Stiftung.
References
- Bagai S., Puri,A., Blumenthal,R. and Sarkar,D.P. (1993) Hemagglutinin-neuraminidase enhances F protein-mediated membrane fusion of reconstituted Sendai virus envelopes with cells. J. Virol., 67, 3312–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K.A., Dutch,R.E., Lamb,R.A. and Jardetzky,T.S. (1999) Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell, 3, 309–319. [DOI] [PubMed] [Google Scholar]
- Baumeister W. and Steven,A.C. (2000) Macromolecular electron microscopy in the era of structural genomics. Trends Biochem. Sci., 25, 624–631. [DOI] [PubMed] [Google Scholar]
- Ben-Efraim I., Kliger,Y., Hermesh,C. and Shai,Y. (1999) Membrane-induced step in the activation of Sendai virus fusion protein. J. Mol. Biol., 285, 609–625. [DOI] [PubMed] [Google Scholar]
- Blumenthal R., Bali-Puri,A., Walter,A., Covell,D. and Eidelman,O. (1987) pH-dependent fusion of vesicular stomatitis virus with Vero cells. Measurement by dequenching of octadecyl rhodamine fluorescence. J. Biol. Chem., 262, 13614–13619. [PubMed] [Google Scholar]
- Böttcher C., Ludwig,K., Herrmann,A., van Heel,M. and Stark,H (1999) Structure of influenza hemagglutinin at neutral and at fusogenic pH by electron cryo-microscopy. FEBS Lett., 463, 255–259. [DOI] [PubMed] [Google Scholar]
- Caffrey M., Cai,M., Kaufman,J., Stahl,S.J., Wingfield,P.T., Covell,D.G., Gronenborn,A.M. and Clore,G.M. (1998) Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J., 17, 4572–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calder L.J., González-Reyes,L., Carcia-Barreno,B., Wharton,S.A., Skehel,J.J., Wiley,D.C. and Melero,J.A. (2000) Electron microscopy of the human respiratory syncytial virus fusion protein and complexes that it forms with monoclonal antibodies. Virology, 271, 122–131. [DOI] [PubMed] [Google Scholar]
- Chen L., Gorman,J.L., McKimm-Breschkin,J., Lawrence,L.J., Tulloch,P.A., Smith,B.J., Colman,P.M. and Larence,M.C. (2001a) The structure of the fusion glycoprotein of Newcastle Disease virus suggest a novel paragdigm for the molecular mehanism of membrane fusion. Structure, 9, 255–266. [DOI] [PubMed] [Google Scholar]
- Chen L., Colma,P.M., Cosgrove,L.J., Lawrence,M.C., Lawrence,L.J., Tulloch,P.A. and Gorman,J.F. (2001b) Cloning, expression, and crystallization of the fusion protein of newcastle disease virus. Virology 290, 290–299. [DOI] [PubMed] [Google Scholar]
- De Carlo S., El-Bez,C., Alvarez-Rua,C., Borge,J. and Dubochet,J. (2002) Cryo-negative staining reduces electron-beam sensitivity of vitrified biological particles. J. Struct. Biol., 138, 216–226. [DOI] [PubMed] [Google Scholar]
- Dutch R.E., Hagglund,R.N., Nagel,M.A., Paterson,R.G. and Lamb,R.A. (2001) Paramyxovirus fusion (F) protein: a conformational change on cleavage activation. Virology, 281, 138–150. [DOI] [PubMed] [Google Scholar]
- Gething M.J., White,J.M. and Waterfield,M.D. (1978) Purification of the fusion protein of Sendai virus: analysis of the NH2-terminal sequence generated during precursor activation. Proc. Natl Acad. Sci. USA, 75, 2737–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh J.K. and Shai,Y. (1999) Direct evidence that the N-terminal heptad repeat of Sendai virus fusion protein participates in membrane fusion. J. Mol. Biol., 292, 531–546. [DOI] [PubMed] [Google Scholar]
- González-Reyes L., Ruiz-Arguello,M.B., Garcia-Barreno,B., Calder,L., López,J., Albar,J.P. Skehel,J., Wiley,D.C. and Melero,J.A. (2001) Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc. Natl Acad. Sci. USA, 98, 9859–9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood A.M. (1974) Characteristics of Sendai virus receptors in a model membrane. J. Mol. Biol., 83, 427–436. [DOI] [PubMed] [Google Scholar]
- Hoekstra D., de Boer,T., Klappe,K. and Wilschut,J. (1984) Fluorescence method for measuring the kinetics of fusion between biological membranes. Biochemistry, 23, 5675–5681. [DOI] [PubMed] [Google Scholar]
- Hsu M., Scheid,A. and Choppin,P.W. (1981) Activation of the Sendai virus fusion protein (F) involves a conformational change with exposure of a new hydrophobic region. J. Biol. Chem., 256, 3557–3563. [PubMed] [Google Scholar]
- Joshi S.B., Dutch,E.E. and Lamb,R.A. (1998) A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology, 248, 20–34. [DOI] [PubMed] [Google Scholar]
- Kohama T., Garten,W. and Klenk,H.-D. (1981) Changes in conformation and change paralleling proteolytic activiation of Newcastle disease virus glycoproteins. Virology 111, 364–376. [DOI] [PubMed] [Google Scholar]
- Lamb R.A. (1993) Paramyxovirus fusion: a hypothesis for changes. Virology, 197, 1–11. [DOI] [PubMed] [Google Scholar]
- Lamb R.A. and Kolakofsky,D. (2001) Paramyxoviridae: the viruses and their replication. In Knipe,D.M. and Howley,P.M. (eds), Fields Virology, 4th edition. Lippincott, Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- Lescar J., Roussel,A., Wien,M.W., Navaza,J., Fuller,S.D., Wengler,G., Wengler,G. and Rey,F.A. (2001) The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell, 105, 137–148. [DOI] [PubMed] [Google Scholar]
- Mancini E.J., Clarke,M., Gowen,B.E., Rutten,T. and Fuller,S.D. (2000) Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol. Cell, 5, 255–266. [DOI] [PubMed] [Google Scholar]
- Melikyan G.B., Markosyan,R.M., Hemmati,H., Delmedico,M.K., Lambert,D.M. and Cohen,F.S. (2000) Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol., 151, 413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P., Pomorski,T., Porwoll,S., Tauber,R. and Herrmann,A. (1996) Transverse movement of spin-labeled phospholipids in the plasma membrane of a hepatocytic cell line (HepG2)—implications for biliary lipid secretion. Hepatology, 24, 1497–1503. [DOI] [PubMed] [Google Scholar]
- Novick S.L. and Hoekstra,D. (1988) Membrane penetration of Sendai virus glycoproteins during the early stages of fusion with liposomes as determined by hydrophobic photoaffinity labeling. Proc. Natl Acad. Sci. USA, 85, 7433–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova E.V., Dube,P., Harris,J.R., Beckmann,E., Zemlin,F., Markl,J. and van Heel,M. (1997) Structure of keyhole limpet hemocyanin Type 1 (KLH1) at 15Å resolution by electron cryomicroscopy and angular reconstitution. J. Mol. Biol., 271, 417–437. [DOI] [PubMed] [Google Scholar]
- Peisajovich S.G., Samuel,O. and Shai,Y. (2000) Paramyxoviruse F1 protein has two fusion peptides: implications for the mechanism of membrane fusion. J. Mol. Biol., 296, 1353–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D., Ovadia,M. and Shai,Y. (1995) A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. EMBO J., 14, 5524–5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey F.A., Heinz,F.X., Mandl,C., Kunz,C. and Harrison,S.C. (1995) The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature, 375, 291–298. [DOI] [PubMed] [Google Scholar]
- Rosenthal P.B., Zhang,X.D., Formanowski,F., Fitz,W., Wong,C.H., MeierEwert,H., Skehel,J.J. and Wiley,D.C. (1998) Structure of the hemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature, 396, 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Argüello B., González-Reyes,L., Calder,L., Palomo,C., Martin,D., Saìz,M.J., Garcia-Barreno,B., Skehel,J.J. and Melero,J.A. (2002) Effect of proteolytic processing at two distinct sites on shape and aggregation of an anchorless fusion protein of human respiratory syncytial virus and fate of the intervening segment. Virology, 298, 317–326. [DOI] [PubMed] [Google Scholar]
- Russell R., Paterson,RG. and Lamb,R.A. (1994) Studies with cross-linking reagents on the oligomeric form of the paramyxovirus fusion protein. Virology, 199, 160–168. [DOI] [PubMed] [Google Scholar]
- Russell C.J., Jardetzky,T.S. and Lamb,R.A. (2001) Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J., 20, 4024–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid A. and Choppin,P.W. (1974) Identification of the biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis and infectivity by proteolytic cleavage of an inactive precursor protein of Sendai virus. Virology, 50, 475–490. [DOI] [PubMed] [Google Scholar]
- Scheid A. and Choppin,P.W. (1977) Two disulfide-linked polypeptide chains constitute the active F protein of paramyxoviruses. Virology, 80, 54–60. [DOI] [PubMed] [Google Scholar]
- Scheid A., Caliguri,L.A., Compans,R.W. and Choppin,P.W. (1972) Isolation of paramyxovirus glycoproteins. Association of hemagglutinating and neuraminidase activities with larger SV5 glycoprotein. Virology, 50, 640. [DOI] [PubMed] [Google Scholar]
- Singh M., Berger,B. and Kim,S. (1999) LearnCoil-VMF: computational evidence for coiled-coil-like motifs in many viral membrane-fusion proteins. J. Mol. Biol., 290, 1031–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi M. and Loyter,A. (1981) Selective extraction of biologically active F-glycoprotein from dithiothreitol reduced Sendai virus particles. FEBS Lett., 131, 381–385. [DOI] [PubMed] [Google Scholar]
- Umino Y., Kohama,T., Sato,T.A., Sugiura,A., Klenk,H.-D. and Rott,R. (1990) Monoclonal antibodies to three structural proteins of Newcastle disease virus: biological characterization with particular reference to the conformational change of envelope glycoproteins associated with proteolytic cleavage. J. Gen. Virol., 71, 1189–1197. [DOI] [PubMed] [Google Scholar]
- van Heel M. (1987) Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy, 21, 111–124. [DOI] [PubMed] [Google Scholar]
- van Heel M. and Harauz,G. (1986) Resolution criteria for three dimensional reconstructions. Optik, 73, 119–122. [Google Scholar]
- Wharton S.A., Skehel,J.J. and Wiley,D.C. (2000) Temperature dependence of fusion by Sendai virus. Virology, 271, 71–78. [DOI] [PubMed] [Google Scholar]
- Wilson I.A., Skehel,J.J. and Wiley,D.C. (1981) Structure of the hemagglutinin membrane glycoprotein of influenca virus at 3 Å resolution. Nature, 289, 366–373. [DOI] [PubMed] [Google Scholar]
- Wriggers W. and Birmanns,S. (2001) Using situs for flexible and rigid-body fitting of multiresolution single-molecule data. J. Struct. Biol., 133, 193–202. [DOI] [PubMed] [Google Scholar]
- Zhao X., Singh,M., Malashkevich,V.N. and Kim,P.S. (2000) Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl Acad. Sci. USA, 97, 14172–14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z.H., Baker,M.L., Jiang,W., Dougherty,M., Jakana,J., Dong,G., Lu,G. and Chiu,W. (2001) Electron cryomicroscopy and bioinformatics suggest protein fold models for rice dwarf virus. Nat. Struct. Biol., 8, 868–873. [DOI] [PubMed] [Google Scholar]