Abstract
A PCR-based method and a reverse transcriptase PCR (RT-PCR)-based method were developed for the detection of pathogenic bacteria in organic waste, using Salmonella spp., Listeria monocytogenes, Yersinia enterocolitica, and Staphylococcus aureus as model organisms. In seeded organic waste samples, detection limits of less than 10 cells per g of organic waste were achieved after one-step enrichment of bacteria, isolation, and purification of DNA or RNA before PCR or RT-PCR amplification. To test the reproducibility and reliability of the newly developed methods, 46 unseeded samples were collected from diverse aerobic (composting) facilities and anaerobic digestors and analyzed by both culture-based classical and newly developed PCR-based procedures. No false-positive but some false-negative results were generated by the PCR- or RT-PCR-based methods after one-step enrichment when compared to the classical detection methods. The results indicated that the level of activity of the tested bacteria in unseeded samples was very low compared to that of freshly inoculated cells, preventing samples from reaching the cell density required for PCR-based detection after one-step enrichment. However, for Salmonella spp., a distinct PCR product could be obtained for all 22 nonamended samples that tested positive for Salmonella spp. by the classical detection procedure when a selective two-step enrichment (20 h in peptone water at 37°C and 24 h in Rappaport Vassiliadis medium at 43°C) was performed prior to nucleic acid extraction and PCR. Hence, the classical procedure was shortened, since cell plating and further differentiation of isolated colonies can be omitted, substituted for by highly sensitive and reliable detection based on nucleic acid extraction and PCR. Similarly, 2 of the 22 samples in which Salmonella spp. were detected also tested positive for Listeria monocytogenes according to a two-step enrichment procedure followed by PCR, compared to 3 samples that tested positive when classical isolation procedures were followed. The study shows that selective two-step enrichment is useful when very low numbers of bacterial pathogens must be detected in organic waste materials, such as biosolids. There were no false-positive results derived from DNA of dead cells in the waste sample, suggesting that it is not necessary to perform RT-PCR analyses when PCR is combined with selective enrichment. Large numbers of added nontarget bacteria did not affect detection of Salmonella spp., L. monocytogenes, and Y. enterocolitica but increased the detection limit of Staphylococcus aureus from <10 to 104 CFU/g of organic waste. Overall, the detection methods developed using seeded organic waste samples from one waste treatment facility (WTF) needed to be modified for satisfactory detection of pathogens in samples from other WTFs, emphasizing the need for extensive field testing of laboratory-derived PCR protocols. A survey of 13 WTFs in Germany revealed that all facilities complied with the German Biowaste Ordinance, which mandates that the end product after anaerobic digestion or aerobic composting be free of Salmonella. In addition, all biosolids were free of L. monocytogenes, Staphylococcus aureus, and Y. enterocolitica, as evidenced by both classical and PCR-based detection methods.
Biological waste can be converted by means of aerobic or anaerobic digestion into biosolids that are usable as soil conditioners and fertilizers in gardening, agriculture, and landscaping. For environmental and health reasons, the product should be tested for the absence of pathogenic microorganisms before its application. As far as pathogenic bacteria are considered, according to German legislation, biosolids have to be free of the indicator organism Salmonella spp., as verified by a classical enrichment procedure (16). However, numerous other pathogenic bacteria are potentially associated with biological waste (54, 59), including Listeria monocytogenes, Yersinia enterocolitica, and Staphylococcus aureus. Classical detection methods are time and labor intensive, since they are usually based on several enrichment steps and subsequent biochemical and serological identification. For this reason, there is a need for alternative, more modern detection methods that are more efficient and faster to perform.
PCR (48) is a powerful tool that allows the species-specific detection of organisms based on nucleic acid amplification. It has been used not only for the identification of isolated bacteria but also for analysis of food (10, 17, 35, 46), clinical (61), and environmental (57) samples. While it is easy to amplify DNA derived from pure cultures, problems arise if the sample investigated is as complex as food, soil, or biological waste, since the PCR is easily inhibited by numerous substances, including humic acids, fats, and proteins (47, 62). Therefore, DNA has to be isolated and purified efficiently, and any PCR-based procedure has to be critically evaluated for its detection limit and reliability. Furthermore, it has to be considered that when using PCR, the DNA of both viable and nonviable cells is amplified. This problem can be overcome either by using an enrichment step before nucleic acid extraction (7, 23) or by performing reverse transcriptase PCR (RT-PCR) to amplify only specific mRNA (28).
This work aimed at developing a PCR- and RT-PCR-based procedure for sensitive, reproducible detection of active bacteria in organic waste samples, which is suited for routine analysis. The research goal was to test the suitability of the PCR for detection of pathogenic bacteria in biological waste, using Salmonella spp. (12), L. monocytogenes (14), Y. enterocolitica (27), and S. aureus (4) as model organisms. At first, detection procedures were developed using seeded samples. To verify the sensitivity and reproducibility of those newly developed methods, nonseeded organic waste samples before and after treatment were collected from diverse aerobic (composting) facilities and anaerobic digestors. They were analyzed by the PCR- and RT-PCR-based methods, as well as the classical detection methods, and the results obtained were compared directly.
MATERIALS AND METHODS
Organic waste samples.
For method development, both freshly suspended waste and anaerobically digested organic waste were used for inoculation experiments. The organic waste was obtained from Kaufbeuren, Germany, and contained biological waste from households with a total solids content of approximately 10% (wt/vol). Samples that were free of the target bacteria were inoculated with the corresponding bacteria as described below.
For validation of the method, 46 samples were collected from diverse aerobic (composting) facilities and anaerobic digestors. The samples consisted of either fresh biological waste from households, which is used as input material for treatment facilities, or of aerobically or anaerobically treated organic waste collected from various stages of the process. To protect the facility owners' identity, the origin of the samples and the type of treatment process were kept anonymous as a precondition to obtaining access to facilities. The total solids content of samples varied from 1.3 to 45.9%.
Bacterial species and inoculation of the organic waste.
Salmonella enterica serovar Typhimurium strain 96 BR 385, Salmonella enterica serovar Senftenberg strain DSM 10062, L. monocytogenes strain I HE/92/1104/666-2 serovar 1/2a, S. aureus strain DSM 3464, and Y. enterocolitica strain DSM 11503 were used as seed organisms. All strains except Y. enterocolitica DSM 11503, which was grown at 30°C, were cultivated in sterile Luria-Bertani (LB) broth at 37°C. Cultures were collected after overnight enrichment. The strains were washed twice and resuspended in 0.9% NaCl solution. Cell numbers were determined by using a Neubauer counting chamber and by plate counting. For inoculation, organic waste samples were used that did not contain the bacterial species to be analyzed, as confirmed by classical isolation methods. One-milliliter portions of dilutions containing 10 to 109 cells were added to 9-ml portions of organic waste, and the preparations were mixed by vortexing. Unseeded waste was used as a control. Experiments were carried out either with freshly seeded organic waste or with samples previously kept for 1 week at 4°C or at room temperature, in order to simulate environmental conditions. Where needed, pasteurization of samples prior to inoculation was achieved by heating to 75°C for 30 min.
Detection of pathogenic bacteria by classical isolation procedures.
The isolation procedure for Salmonella spp. was performed essentially according to Appendix 2 of the German Biowaste Ordinance (16). For isolation of Salmonella spp., suspended organic waste was diluted 1:10 in peptone water and incubated at 37°C for 20 h. Aliquots of 0.1 ml of this broth were then diluted in 10 ml of Rappaport Vassiliadis medium (Merck) and kept at 37 and 43°C for 24 h (58). Isolation of bacteria was carried out on xylose lysine deoxychlorate plates (Merck) by incubation at 37°C for 24 h (13).
In the case of other pathogens, proven isolation techniques reported in the literature were assessed by testing them on seeded biosolid samples. In general, one to two selective enrichment steps in broth were made before plating on solid selective media. For L. monocytogenes, 1 g of waste sample was inoculated in 9 ml of UVM-I broth (Merck) and kept at 30°C for 24 h (45). Next, 0.1 ml was transferred into 10 ml of Fraser broth (Merck) and incubated for 24 h at 30°C (37). Cells were plated on Oxford selective agar (Merck) and incubated for 48 h at 37°C (11). L. monocytogenes formed gray colonies with a black diffusion zone. For isolation of Y. enterocolitica, 1 g of waste sample was inoculated into 9 ml of Yersinia selective enrichment broth according to Ossmer and incubated for 24 h at 30°C (49) The suspension was streaked on CIN Agar (Merck) and kept another 24 h at 30°C (1, 49). To detect S. aureus, 1 g of biological waste was mixed with 9 ml of Giolitti-Cantoni medium (Merck) containing potassium tellurite as selective reagent and incubated anaerobically (with a layer of sterile paraffin) for 24 h at 37°C (18). The suspension was streaked on Baird Parker (3) agar plates (Merck) and kept at 37°C for 48 h (2). Presumptive colonies were confirmed by PCR analysis as described below. For identification of Salmonella spp., the Wellcolex color Salmonella test (Abbott) was used as described by the manufacturer.
Confirmation of presumptive colonies by PCR.
Presumptive colonies were picked from the agar plate, suspended in LB broth, and incubated for about 1 h in a shaking incubator at 37°C (Salmonella spp., L. monocytogenes, and S. aureus) or at 30°C (Y. enterocolitica). Two microliters of cell culture was added to the PCR mixture as a template.
Enrichment of samples prior to nucleic acid extraction.
Before extraction of DNA or RNA, biological waste was suspended in enrichment broth at a ratio of 1:10. When inoculation experiments were performed, usually 1 g of waste samples was mixed with 9 ml of enrichment broth, so the term “detection limits” refers to 1 g of organic waste. When unseeded samples were tested, usually 10 g of waste was diluted in 90 ml of enrichment broth. One-step enrichment was performed as indicated in Table 1, depending on the target bacterium. Enrichment broth, incubation time, and temperature for optimal detection were determined in preliminary experiments with inoculated samples.
TABLE 1.
Enrichment procedures prior to nucleic acid extraction and reference strains used
| Target bacterium | Reference strain | Enrichment broth | Incubation
|
|
|---|---|---|---|---|
| Temp (°C) | Length (h) | |||
| Salmonella spp. | S. enterica serovar Typhimurium 96 BR 385 | Peptone water | 37 | 20 |
| S. enterica serovar Senftenberg DSM 10062 | ||||
| L. monocytogenes | I HE/92/1104/666-2 serovar 1/2a | UVM-1 medium | 30 | 24 |
| Y. enterocolitica | 11503 | Ossmer broth | 30 | 24 |
| DSM 4780 | ||||
| DSM 9499 | ||||
| DSM 1502 | ||||
| S. aureus | DSM 3464 | Giolitti-Cantoni brotha | 37 | 24 |
Broth was overlayed with sterile paraffin; no potassium tellurite was added.
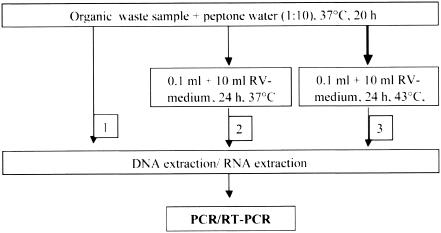
For L. monocytogenes and Y. enterocolitica, selective enrichment turned out to be advantageous, while Salmonella spp. was enriched in nonspecific peptone water leading to sensitive detection (7). For enrichment of S. aureus, no potassium tellurite was added as a selective reagent, since its addition led to markedly higher detection limits (104 instead of 10 cells per g), indicating inhibition of the target organism itself. For detection of Salmonella spp. in nonamended samples, a two-step enrichment was performed with Rappaport Vassiliadis medium (58) as the second, selective enrichment broth prior to nucleic acid extraction and PCR (Fig. 1). Similarly, for detection of L. monocytogenes in unseeded samples, nucleic acid extraction was performed after one-step enrichment and after a second enrichment step in Fraser bouillon (Fig. 2).
FIG. 1.
Flowchart for the detection of Salmonella spp. using different methods of enrichment prior to nucleic acid extraction and PCR.
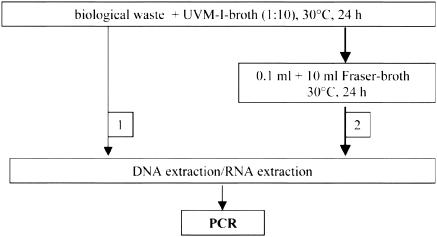
FIG. 2.
Flowchart for the detection of Listeria monocytogenes using different methods of enrichment prior to nucleic acid extraction and PCR.
DNA isolation.
The extraction procedure was performed essentially as described before (7) with slight modifications. After one-step enrichment, the mixture was centrifuged at 200 × g for 5 min to allow bigger particles to settle (This step can be omitted after the second enrichment step.) One milliliter of the supernatant containing bacterial cells was transferred into a 2-ml Eppendorf cap, and cells were pelleted at 20,800 × g for 5 min in a cooled microcentrifuge (4°C). After careful removal of the supernatant, the pellet was suspended in 400 μl of lysis buffer (0.5% N-laurylsarcosine, 50 mM Tris-Cl, 25 mM EDTA [pH 8.0]), containing freshly prepared glycogen (0.03 μg/μl). One microliter of proteinase K (20 mg/ml) was added to the suspension, and the mixture was incubated for 1 h at 37°C. After incubation, 600 μl of an NaI solution (6 M NaI in 50 mM Tris-Cl, 25 mM EDTA [pH 8.0]) and 1 ml of isopropanol were added, and the mixture was centrifuged at 20,800 × g for 5 min. The pellet was washed with 35% isopropanol and resuspended in 100 μl of sterilized water by pipetting.
Crude DNA extracts were purified with the Wizard PCR Preps DNA purification system (Promega, Madison, Wis.) as described by the manufacturer by using 2-ml syringes to pass the extract through the minicolumn. The DNA was eluted by using 50 μl of hot (80°C) TE buffer (10 mM Tris-Cl, 1 mM EDTA [pH 8.0]). For detection of Salmonella spp., S. aureus, and Y. enterocolitica, the extract was purified over one minicolumn. For successful amplification of DNA from organic waste seeded with L. monocytogenes, it was necessary to pass the extract through a second minicolumn.
RNA isolation.
RNA extraction was performed after enrichment with the RNeasy-Mini kit (Qiagen). After incubation, the enrichment broth was centrifuged at 200 × g for 5 min, and 1 ml of supernatant containing bacterial cells was transferred into an RNase-free 1.5-ml Eppendorf cap (BioPur; Eppendorf) and centrifuged at maximum speed for 5 min. After carefully removing the supernatant, 100 μl of TE buffer containing 6 μl of a 50-mg/ml lysozyme stock solution (lysozyme stock solution stored in single-use aliquots) was added to the pellet, mixed by pipetting, and incubated for 15 min at room temperature. After adding 350 μl of RLT buffer (kit component) containing 10 μl of β-mercaptoethanol per ml of RLT and vigorously vortexing the mixture, the solution was centrifuged at maximum speed for 2 min. For the following steps, only the nucleic acids-containing supernatant was used, which was purified as described by the manufacturer. RNA was eluted from the column with 50 μl of RNase-free water.
Since the RNA extract was still contaminated with amounts of DNA that can be coamplified by the RT-PCR, a DNA digestion was performed. Ten microliters of the extracted RNA-solution was transferred into a 500-μl RNase-free Eppendorf cap, 3 μl of RQ1 RNase-free DNase (Promega) was added (1 U/μl), and the resulting solution was mixed carefully. The DNA was digested by incubating the cap at 37°C for 30 min. For inactivation of the DNase, the solution was heated to 75°C for 5 min and subsequently cooled on ice. The resulting solution was either immediately used for RT-PCR or frozen at minus 80°C. Instructions concerning the handling and storage of RNA are given in Appendix A of the RNeasy Mini Handbook (Qiagen).
Selection of primers.
Primers utilized for PCR and RT-PCR are indicated in Table 2. They were selected after a literature research and extensive testing on pure cultures. In addition, databases available at the National Center for Biotechnology Information were searched by BLAST to exclude matches with other known sequences. The primer pairs were selected not only for specificity of the target organism, but also for their suitability for mRNA detection by RT-PCR. For Salmonella spp., a primer pair was chosen that amplifies a 159-bp-long fragment within the ompC gene. It has been described as being specific for Salmonella spp. (31) and being constitutively expressed along the growth curve at high levels at both low and high osmolarities (36), which makes it suitable as well for mRNA detection. A primer pair amplifying a species-specific sequence within the iap gene (6) was selected for detection of L. monocytogenes. Klein et al. (28) described this gene as a good target for specific detection of viable L. monocytogenes based on RT-PCR amplification. For detection of S. aureus, the nuc gene encoding the thermostable nuclease (5) was selected as the target gene for both PCR and RT-PCR. No literature about mRNA expression of this gene was available, but preliminary experiments showed easily detectable RT-PCR products when pure cultures were used. Primer pairs for genes that were described as suitable for detection of pathogenic Y. enterocolitica did not amplify DNA derived from four available strains of Y. enterocolitica (DSM 4780, DSM 9499, DSM 1502, and DSM 11503). Therefore, a primer pair specifically binding to a 16S ribosomal DNA (rDNA) sequence of Y. enterocolitica (55) was selected for Y. enterocolitica-specific PCR and RT-PCR. Hence, in this case, rRNA and not mRNA was amplified by RT-PCR.
TABLE 2.
Primers for DNA and RNA amplification
| Target bacterium (target gene) | Oligonucleotide sequences (5′→3′) | Amplification region (bp) | Annealing temp (°C) | Reference |
|---|---|---|---|---|
| Salmonella spp. (ompC) | S18: ACCGCTAACGCTCGCCTGTAT | 159 | 56 | 31 |
| S19: AGAGGTGGACGGGTTGCTGCCGTT | ||||
| L. monocytogenes (iap) | ELMIAPF: CAAACTGCTAACACAGCTACT | 371 | 60 | 28 |
| ELMIAPR: GCACTTGAATTGCTGTTATTG | ||||
| S. aureus (nuc) | nuc1: GCGATTGATGGTGATACGGTT | 270 | 55 | 5 |
| nuc2: AGCCAAGCCTTGACGAACTAAAGC | ||||
| Y. enterocolitica (16S rDNA)a | Y.16S-86f: GCGGCAGCGGGAAGTAGTTTA | 416 | 53 | 55 |
| Y.e.eur. 16S-455r: CAATCACAAAGGTTATTAACCTTTATG |
rDNA, ribosomal DNA.
PCR amplification conditions.
PCR was performed in 100-μl reaction mixtures using the HotStar Taq DNA polymerase (Qiagen). The PCR mixture contained 10 μl of 10× PCR buffer (containing 15 mM MgCl2), 2 μl of deoxynucleoside triphosphate (dNTP) mix (containing 10 mM each dNTP), 0.5 μl of each primer solution (100 μM), 0.5 μl of the HotStar Taq DNA polymerase (5 U/μl), the template DNA (2 μl of the DNA extract), and deionized water for a final volume of 100 μl. The reaction mixture was subjected to PCR under the following conditions: an initial step for activation of the HotStar Taq DNA polymerase at 95°C for 15 min followed by 35 cycles of denaturation at 94°C for 45 s, primer annealing for 1 min (annealing temperatures are indicated in Table 2), and DNA extension at 72°C for 1 min. After the last amplification cycle, samples were kept for final extension at 72°C for 10 min and immediately cooled to 4°C.
RT-PCR amplification conditions.
For RT-PCR, both the rTth DNA polymerase (Perkin-Elmer) and the Qiagen one-step RT-PCR kit were applied to compare the efficiencies of the two systems. The reactions were done according to the manufacturers′ instructions. When the rTth DNA polymerase was used, the reaction was performed in a total volume of 100 μl. First, 20 μl of a mix was prepared for reverse transcription, containing 1× rTth RT buffer, 1 mM MnCl2, 200 μM each dNTP, the downstream primer (0.75 μM), the rTth DNA polymerase (5 U), and 2 μl of the RNA extract, and then the mixture was subjected to 60°C for 35 min. In the presence of MnCl2, the rTth DNA polymerase reverse transcribes RNA at elevated temperatures. Subsequently, the assay was cooled on ice, and 80 μl of a mixture for DNA amplification containing 8 μl of chelating buffer, 6 μl of MgCl2 solution (25 mM), 1 μl of the upstream primer solution (100 pM), and 65 μl of water was added. The final concentration of the components in the combined RT-PCR-PCR mix was 0.8× chelating buffer, 15 mM MgCl2, and 0.15 μM upstream primer. DNA polymerase activity is enhanced by chelating Mn2+ and adding MgCl2. The resulting mixture was subjected to the corresponding PCR temperature cycles.
When the Qiagen one-step RT-PCR kit was used, the volume of the total reaction mixture was 50 μl. The mixture contained 10 μl of 5× Qiagen one-step RT-PCR buffer (including 12.5 mM MgCl2), 2 μl of dNTP mix (containing 10 mM each dNTP), 0.6 μM each primer, 2 μl of Qiagen one-step RT-PCR enzyme mix (containing the two RTs Omniscript and Sensiscript and the HotStar Taq DNA polymerase), the template RNA (2 μl of the RNA extract), and deionized water for a final volume of 50 μl. The mixture was subjected to RT-PCR without the need to open the vials during the reaction. For reverse transcription, the vial was kept at 50°C for 30 min. Subsequently, the temperature was raised to 95°C for 15 min in order to activate the HotStar TaqDNA polymerase. At the same time, RTs were denatured. From then onwards, PCR amplification was performed with the corresponding PCR program as described above.
Gel electrophoresis.
PCR products were identified by estimating sizes after agarose gel electrophoresis on a 1.3% agarose gel (100 mA/80 V) and staining with ethidium bromide.
RESULTS
Detection limits achieved by DNA extraction and PCR using inoculated waste samples.
DNA extraction and PCR detection were performed after various steps to assess the reliability of the detection procedure and simulate environmental conditions. Detection limits were less than 10 cells per g of waste sample for all bacterial strains tested (Table 3) when an enrichment step was performed prior to DNA extraction (Table 1). The number of bacterial cells in the mixture of waste sample and enrichment broth (1:10) needed for positive PCR detection in the absence of incubation was 106/ml, corresponding to 107/g of organic waste for all target organisms except S. aureus, for which it was 1 order of magnitude less (Table 3). Detection limits in inoculated waste samples that had previously been pasteurized were similar to those in samples that were just suspended but not incubated in the enrichment broth (Table 3). For this experiment, waste samples were spiked and pasteurized as described in Materials and Methods. Next they were subjected to the complete detection procedure, including one-step enrichment, DNA extraction, and PCR. Due to the enrichment step, dead cells were excluded from detection at concentrations below 107 cells (Salmonella spp., L. monocytogenes, and Y. enterocolitica) or 106 cells (S. aureus) per g of waste sample.
TABLE 3.
Detection limits of bacteria tested by DNA extraction and PCR in inoculated organic waste samples
| Target organism | PCR detection limit (CFU/g of waste sample)a
|
|||||
|---|---|---|---|---|---|---|
| After enrichmentb | Without enrichmentc | Pasteurized samplesb,d | After 1 wk of incubation at:
|
In the presence of nontarget bacteriab,e | ||
| Room temp | 4°C | |||||
| Salmonella spp. | <10 | 107 | 107 | 10 | 10 | 10 |
| L. monocytogenes | <10 | 107 | 107 | 102 | 102 | 10 |
| Y. enterocolitica | <10 | 107 | 107 | 102 | 103 | 10 |
| S. aureus | <10 | 106 | 106 | 104 | 104 | 104 |
This specification refers to 1 g of waste being suspended in enrichment broth.
Enrichment was performed as indicated in Table 1.
Waste samples were suspended in enrichment medium and extracted immediately.
Samples were heated to 75°C for 30 min before enrichment.
The sample was additionally seeded with a large number (107/g) of each of the other three bacterial strains tested that were not target organisms in the experiment.
To simulate natural conditions, inoculated waste samples were kept for 1 week at room temperature or 4°C. In the case of Salmonella, detection limits remained at 10 originally inoculated cells per g of waste, whereas detection limits for the other bacteria tested changed (Table 3). The reason for this change is most likely a loss of cell viability during storage. Samples were also inoculated with a large number (107/g) of the other three strains of bacteria tested in addition to the target organism. For Salmonella spp., L. monocytogenes, and Y. enterocolitica, detection limits were not affected. However, the detection limit of S. aureus changed markedly in the presence of large numbers of nontarget bacteria (Table 3). This result may be due to the loss of selectivity of the enrichment medium by omitting potassium tellurite.
Detection limits achieved by RNA extraction and RT-PCR using inoculated waste samples.
For detection of RNA, inoculated samples were enriched under the same conditions used for DNA extraction (Table 1). The RT-PCR detection limits achieved by one-step enrichment, RNA extraction, and RT-PCR with either rTth DNA polymerase (Perkin-Elmer) or the Qiagen one-step RT-PCR kit were as follows. Using rTth DNA polymerase, no reproducible RT-PCR products could be detected unless a nested RT-PCR was performed (results not shown). However, the reproducibility of RT-PCR results was good if the same RNA extract was amplified by the Qiagen one-step RT-PCR kit, with a detection limit of <10 CFU per g of waste for Salmonella spp., L. monocytogenes, and Y. enterocolitica. For S. aureus, the detection limit was 104 CFU/g of waste. These results indicate that the RNA extracts derived from organic waste samples used in our experiments were amplified much more efficiently by the Qiagen one-step RT-PCR kit. This observation shows that it is advisable to test different enzyme mixtures for amplification of nucleic acid extracts derived from the same samples. The Qiagen one-step RT-PCR kit was used for all further analyses.
Since amplification of the RNA extracts by PCR without prior reverse transcription did not result in PCR products, it was concluded that the RT-PCR products were derived from bacterial RNA and not from DNA. A DNA digestion step prior to amplification was crucial to the procedure.
Detection of pathogenic bacteria by classical isolation methods in unseeded biological waste samples.
For the comparison of the PCR- and RT-PCR-based detection methods with the classical isolation procedures, samples were taken from various organic waste treatment facilities. Salmonella spp. could be detected in 22 of the 46 samples taken by classical isolation techniques (Table 4). Eight of the positive samples had not been treated, and 14 samples were from intermediate stages of a digestor. All of the facilities tested produced biosolids free of Salmonella. Three of the 22 samples were also positive for L. monocytogenes. However, neither Y. enterocolitica nor S. aureus was detected in the 46 waste samples analyzed by classical isolation methods.
TABLE 4.
Pathogen detection in waste treatment facilities by classical isolation procedures and detection of Salmonella spp. after various enrichment procedures followed by DNA extraction and PCR or RNA extraction and RT-PCR
| Waste treatment facility sample (only Salmonella- positive samples [n = 22]) | % Total solids | Detection by classical isolation of:
|
Detection of Salmonella spp. after enrichment in:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptone water
|
Peptone water + RV
|
||||||||||
| 37°C
|
43°C
|
||||||||||
| Salmonella spp. | L. monocytogenes | S. aureus | Y. enterocolitica | PCR | RT-PCR | PCR | RT-PCR | PCR | RT-PCR | ||
| A | 5.4 | + | + | − | − | − | NDa | − | ND | + | ND |
| B | 8.5 | + | − | − | − | − | − | − | − | + | + |
| C | 5.1 | + | − | − | − | + | − | + | + | + | + |
| D | 7.5 | + | − | − | − | + | ND | + | ND | + | ND |
| E | 33.9 | + | − | − | − | − | ND | − | ND | + | ND |
| F | 5.0 | + | − | − | − | − | ND | + | ND | + | ND |
| G | 1.3 | + | − | − | − | + | ND | + | ND | + | ND |
| H | 6.9 | + | − | − | − | − | ND | + | ND | + | ND |
| I | 33.4 | + | − | − | − | + | ND | + | ND | + | ND |
| J | 10.2 | + | + | − | − | − | ND | + | ND | + | ND |
| K | 1.5 | + | − | − | − | − | ND | + | ND | + | ND |
| L | 36.3 | + | − | − | − | − | ND | − | ND | + | ND |
| M | 28.9 | + | + | − | − | − | − | − | + | + | + |
| N | 7.0 | + | − | − | − | − | − | − | − | + | + |
| O | 33.8 | + | − | − | − | − | − | + | + | + | + |
| P | 45.9 | + | − | − | − | − | − | − | − | + | + |
| Q | 29.1 | + | − | − | − | − | − | + | + | + | + |
| R | 29.7 | + | − | − | − | − | − | − | − | + | + |
| S | 5.9 | + | − | − | − | − | ND | − | ND | + | ND |
| T | 1.3 | + | − | − | − | + | ND | + | ND | + | ND |
| U | 34.0 | + | − | − | − | + | ND | + | ND | + | ND |
| V | 5.9 | + | − | − | − | − | ND | + | ND | + | ND |
| Total (no. positive/no. tested) | 22/22 | 3/22 | 0/22 | 0/22 | 6/22 | 0/8 | 13/22 | 4/8 | 22/22 | 8/8 | |
ND, not determined.
Detection of Salmonella spp. in unseeded samples by DNA extraction and PCR.
Each sample taken was analyzed by nucleic acid extraction and PCR. When DNA was extracted after one-step enrichment (in peptone water), a Salmonella-specific PCR product could be obtained only for 6 of the 22 samples that tested positive for Salmonella spp. by the classical isolation method (Table 4). Therefore, the PCR detection method needed to be further optimized. The most effective way of gaining optimal sensitivity was to carry out a further enrichment step before DNA extraction. Hence, DNA extraction was performed after three different enrichment protocols (Fig. 1).
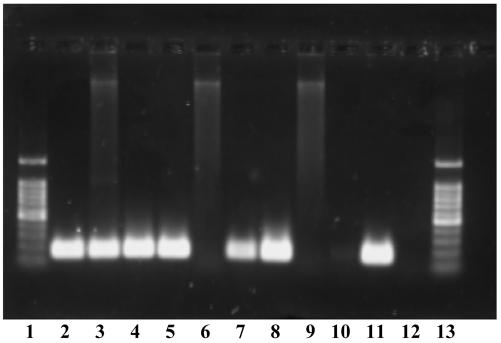
Thirteen of the 22 culture-positive samples tested positive by PCR, when DNA was extracted after enrichment in peptone water and in Rappaport Vassiliadis medium incubated at 37°C. However, after a two-step enrichment in peptone water and Rappaport Vassiliadis medium that had been incubated at 43°C, a temperature that is more selective than 37°C, a PCR product could be obtained for all the 22 culture-positive samples, but for none of the culture-negative samples. Salmonella-specific PCR products obtained from three samples (samples I, J, and M) after three different methods of enrichment are shown on an ethidium bromide-stained agarose gel (Fig. 3). For sample I, it was possible to obtain a PCR product after one-step enrichment, whereas a second enrichment step in Rappaport Vassiliadis medium was necessary for PCR detection in sample J. Similarly, for sample M, a PCR product visible on the agarose gel was generated only after a second enrichment step in Rappaport Vassiliadis medium at 43°C. There was no correlation between total solids content and enrichment procedure necessary to obtain a PCR product (Table 4). Therefore, it can be concluded that only cell number and cell activity were decisive factors for DNA amplification.
FIG. 3.
Agarose gel electrophoresis of PCR-amplified DNA from the ompC gene of Salmonella spp. derived from various biological waste samples under different conditions of enrichment (Fig. 1) and DNA extraction. Lanes: 1, 100-bp ladder as size marker; 2, pure culture of S. enterica serovar Typhimurium as positive control; 3, waste sample I (Table 4) with enrichment in peptone water; 4, waste sample I with enrichment in peptone water and RV medium at 37°C; 5, waste sample I with enrichment in peptone water and RV medium at 43°C; 6, waste sample J with enrichment in peptone water; 7, waste sample J with enrichment in peptone water and RV medium at 37°C; 8, waste sample J with enrichment in peptone water and RV medium at 43°C; 9, waste sample M with enrichment in peptone water; 10, waste sample M with enrichment in peptone water and RV medium at 37°C; 11, waste sample M with enrichment in peptone water and RV medium at 43°C; 12, water as a negative control; 13, 100-bp ladder as size marker.
None of the 24 samples that tested negative for Salmonella spp. by the classical procedure showed PCR amplification of Salmonella-specific DNA regardless of the enrichment protocol used. Hence, there were no false-positive results due to the presence of dead cells, free DNA, or unspecific amplification associated with the procedure developed based on DNA extraction.
Detection of Salmonella spp. in unseeded samples by RNA extraction and RT-PCR.
Eight of the samples that tested positive for Salmonella spp. were also analyzed by RNA extraction and RT-PCR. The enrichment procedures were the same as those used before when extracting DNA (Fig. 1). Here, a two-step enrichment was essential to achieve an RT-PCR product for all samples investigated. After a second step of enrichment in Rappaport Vassiliadis broth at 37°C, only half of the samples showed an amplification product, whereas all of the samples tested positive by RT-PCR after two-step enrichment at 43°C (Table 4).
Detection of L. monocytogenes in unseeded samples by PCR or RT-PCR.
Only 3 of the 46 samples tested positive for L. monocytogenes by classical isolation methods. Isolation of DNA and RNA was performed either after one-step enrichment in UVM-I broth (30°C, 24 h) or after two-step enrichment in UVM-I broth and Fraser broth (30°C, 24 h; Fig. 2).
In none of the three samples was it possible to detect L. monocytogenes after one-step enrichment by DNA extraction and PCR, but in two samples, two-step enrichment resulted in positive detection. Hence, in one of the three samples, no RT-PCR product that would be visible on an agarose electrophoresis gel was obtained after one-step or two-step enrichment.
Detection of Y. enterocolitica and S. aureus in unseeded samples by PCR or RT-PCR.
Neither Y. enterocolitica nor S. aureus was detected in any of the 46 samples by the PCR or RT-PCR procedure. This was consistent with the results obtained by the classical procedure. However, for both organisms, the detection procedure was based on just one-step enrichment.
DISCUSSION
Salmonella spp. and a number of other pathogenic microorganisms are frequent contaminants of biological waste. Within the scope of this study, 48% of the 46 samples investigated tested positive for Salmonella, which could be detected in both untreated and treated waste samples during intermediate stages of treatment. Before applying biological waste to fields or gardens, it has to be certified as being free of pathogenic microorganisms to avoid any possible infection risks for human beings and grazing animals (16). A great deal of time and effort is required to detect food-borne pathogens in biological waste when only classical enrichment methods are considered. Furthermore, the results obtained on selective and differential media are often ambiguous, which necessitates further testing by biochemical and serological tests. For this reason, we evaluated the application of PCR technology to organic waste samples.
Generally, PCR enables a sensitive species-specific detection without the necessity of cell cultivation. However, there are some aspects that have to be considered if PCR is applied for the detection of microorganisms in environmental samples. Complex samples like biological waste contain numerous inhibitory substances (e.g., humic acids) that can inhibit the PCR completely or might lead to high detection limits (47, 62). Frequently, the number of target organisms is small against a background of large numbers of nontarget cells (9). Moreover, DNA is quite stable and might be detected not only as part of living cells but also as part of dead cells or as free DNA (26, 38). Hence, for successful, sensitive PCR detection of viable cells, it is necessary to remove inhibitory substances efficiently, concentrate target organisms (33), and use a system that excludes dead cells from detection. In this study, the combined effect of those three factors could be addressed by including two enrichment steps prior to nucleic acid extraction. However, DNA and RNA extracts still needed to be purified by passing them through commercially available clean-up columns.
Selection of target genes for PCR and RT-PCR detection.
The first step in method development was to select target genes that are suitable for detection of both DNA and mRNA, with the latter indicating cell viability. For successful amplification of mRNA, not only is it necessary to choose a gene that is selective for the target organism, but in addition, the gene should be expressed at a high level at the time RNA is extracted.
The ompC gene of Salmonella enterica serovar Typhi encodes a major outer membrane protein and is found to be highly conserved in different Salmonella serotypes (42). It is, in contrast to Escherichia coli, highly expressed at both high and low osmolarities (36, 44). Its structure is similar to that of the E. coli ompC gene, yet it differs in some regions (43). The primer set used in these investigations was derived from those heterologous regions (31). Preliminary experiments showed that after enrichment in peptone water at 37°C for 20 h, ompC-specific RT amplification of extracted RNA resulted in well-detectable amplification products.
For detection of L. monocytogenes, the iap gene encoding p60, a major extracellular protein that is possibly associated with invasion of nonprofessional phagocytic cells (29), was chosen as a target for PCR and RT-PCR amplification. Although the p60-related proteins occur in all Listeria species, nucleotide sequences of the corresponding iap genes demonstrated common and variable regions within the p60 proteins. On this basis, a specific primer set for detection of L. monocytogenes has been designed (6). Klein and Juneja (28) investigated the detection of viable L. moncytogenes by RT-PCR by amplifying three different genes, including the iap gene by using the primers reported by Bubert et al. (6). Furthermore, the hly gene coding for the 58-kDa virulence factor listeriolysin O (39) and the prfA gene coding for a 27.1-kDa protein that has been shown to positively regulate the expression of several Listeria virulence factors (8) were tested for their suitability as target genes for amplification of specific mRNA. According to the experiments of Klein and Juneja (28), the sensitivity for detection of the iap gene in L. moncytogenes cultures by RT-PCR was markedly higher than those for hly and prfA. Based on these results and successfully performed preliminary experiments with inoculated waste samples, the iap gene was chosen as a target gene for both PCR and RT-PCR detection in this study.
S. aureus DNA and mRNA were amplified with a primer pair targeting the nuc gene, which encodes the thermostable nuclease of S. aureus. Brakstad et al. (5) selected a primer pair on the basis of the published sequence of the 966-bp nuc gene of S. aureus (50). The primers had been successfully tested by the authors with 90 reference or clinical S. aureus strains, whereas 80 other strains, including staphylococcal species producing a thermostable nuclease, were PCR negative. Preliminary experiments conducted in the present study showed that the mRNA expressed by the nuc gene was also a good target for RT-PCR detection after overnight enrichment at 37°C in GC broth, since an easily detectable RT-PCR product was generated. Thus, the nuc gene was selected as a target for PCR and RT-PCR using the primer set published by Brakstad et al. (5).
For Y. enterocolitica, no specific gene suitable for PCR detection was found. There are several PCR systems described in the literature for identification of pathogenic Y. enterocolitica targeting the chromosomally encoded ail gene for attachment and invasion (30, 40) and the yst gene encoding a heat-stable enterotoxin (25). However, although the four strains of Y. enterocolitica tested (DSM 4780, DSM 9499, DSM 11502, and DSM 11503) were clinical isolates, no PCR product could be obtained. Therefore, a primer pair targeting rRNA described by Trebesius et al. (55) was selected. With this primer pair, amplification products were generated from all test strains. Hence, in this case, the more stable rRNA was amplified by RT-PCR.
PCR detection based on DNA extraction.
Several authors directly extracted DNA from complex materials, such as soil (51, 56, 63), sediments (34), or food (35). Drawbacks of direct extraction of DNA include the codetection of dead cells and the fact that due to inhibitory substances, detection limits are usually higher than those achieved by classical isolation methods. In this work, we aimed at developing and field-testing a method that excludes the detection of dead cells and shows the same sensitivity as the classical isolation procedure. A simple means intended to favor the detection of viable cells and, at the same time, lower the detection limit is to include an enrichment step prior to DNA extraction. This approach has been reported mainly for the qualitative detection of pathogens in food (15, 19, 47, 52, 60). In this study, sensitive detection could be achieved with samples containing freshly inoculated cells of Salmonella spp. after enrichment in nonselective peptone water. However, as the experiments with unseeded samples showed, sensitive detection within an environment of target bacteria can be markedly improved if highly selective enrichment is performed that allows only the target bacteria to multiply.
Experiments with seeded samples showed that when one enrichment step was applied before DNA extraction, dead cells were only detected if present at a very high level (107/g), whereas freshly inoculated cells could be detected at levels less than 10 cells per g of waste sample. Hence, when using the procedure mentioned above, dead cells were for the most part excluded from detection.
RT-PCR detection based on RNA extraction.
Another way of detecting only viable cells is the amplification of mRNA, which is present only in active cells. However, when detecting mRNA, there are some aspects to be considered. First, due to the ubiquitous and very stable RNAses, mRNA is quickly degraded unless those enzymes are destroyed or at least inhibited. Second, small amounts of DNA are concurrently extracted and result in an amplification product by RT-PCR, unless they are completely digested by DNases. Otherwise, false-positive results might be generated, possibly derived from DNA of dead cells.
Moreover, the expression of different genes varies strongly, depending on the gene product and on environmental conditions, which makes the selection of suitable primers more difficult. In this study, as in the case of detection of DNA, an enrichment step turned out to be advantageous for the detection of mRNA, due to the fact that the number of target cells and thus of detectable RNA molecules increases markedly. Many genes are expressed at their highest level in active cells, and it was not possible to achieve a sensitive detection based on DNA or RNA extraction without performing an enrichment step. It is interesting to note that one commercial kit gave superior results to another, underscoring the observation by Al-Soud and Rådstrõm that components in biological samples can have different effects on different polymerases in PCR (1).
Discrepancy between unseeded and inoculated samples.
As shown in this study, detection limits were different when biological waste samples inoculated with fresh bacterial cells from a laboratory culture were compared to unseeded samples with target cells showing lower activity. With freshly inoculated samples, detection limits for all bacteria tested were low (<10 per sample) when using the originally developed detection procedure, which is based on single-step enrichment. In contrast, it was not possible to obtain the same sensitivity as the classical detection procedure when analyzing unseeded samples from various waste treatment facilities. This is due to the fact that not only cell number but also cell activity plays an important role, especially when an enrichment step is involved. Both cell activity and parameters such as the presence and activity of target bacteria can alter detection limits dramatically. Those facts have an impact on how fast and to what level bacterial cells grow in an enrichment culture.
Hence, for Salmonella spp. and L. monocytogenes, a two-step enrichment prior to nucleic acid extraction was essential to achieve a sensitivity by PCR analysis equal to that of classical detection. This was the case for both the DNA-based and RNA-based procedures. The fact that neither Y. enterocolitica nor S. aureus was detected in natural organic waste samples might be due to insufficient enrichment, since for detection of those organisms, only one-step enrichment was employed.
These results indicate that both the number and activity of target cells were probably low in most of the organic waste samples tested, differing from laboratory samples inoculated with bacterial cells from fresh cultures. They underscore the importance of subjecting procedures that have been developed under laboratory conditions to critical field conditions using unseeded samples. Otherwise, reliable predictions cannot be made concerning the appropriateness of a newly developed detection procedure. In a study designed to enumerate relative concentrations of pathogens in stormwater drain samples, detection limits were found to be highly dependent on the type of organism tested for (33). Detection limits were calculated as a function of volume of sample processed through filters, recovery of organisms from filters, inhibition of PCR assays, and fraction of sample analyzed by PCR. It was shown that for Giardia lamblia and Shigella, the detection limit was influenced mostly by the sample volume employed, whereas for Salmonella, E. coli O157:H7, and Cryptosporidium parvum, detection was highly affected by inhibitory substances. The detection limits for viruses were influenced by both filter recovery and PCR inhibition. Enrichment procedures were not utilized in that study, and the authors stressed the importance of defining detection limits on a sample-by-sample basis (33). Clearly, environmental matrices present a formidable barrier to PCR-derived pathogen detection methods.
Sensitivity of PCR product detection.
There are several ways to increase the sensitivity of PCR product detection, including nested PCR (60), Southern blotting (53), and quantitative real-time PCR (20).When using nested PCR, the risk of contamination due to DNA aerosols increases strongly, since vials with amplified DNA have to be opened to add reagents needed for a second PCR. Several nested PCR procedures were tested during this research, leading repeatedly to false-positive results. Hence, we do not recommend nested PCR for routine analysis. Southern blotting not only increases the sensitivity of the PCR product detection (28), but also verifies the product when a specific hybridization probe is used. However, Southern blotting adds a significant additional expenditure of time and effort; thus, it is more appropriate for nonrecurring applications to verify ambiguous PCR products than for routine analysis. A very promising approach is the use of real-time PCR (20) based on the fluorogenic 5′-nuclease technique (32), which increases both sensitivity and specificity of product detection due to a specifically binding, labeled probe between the pair of primers. Additionally, amplified DNA molecules can be analyzed not only qualitatively but also quantitatively, although the same restrictions due to matrix-related inhibition of PCR apply. There is no need for gel electrophoresis, since the amount of the PCR product is measured directly by the increase in fluorescence in the vial. These qualities make it very well suited for routine analysis, and several protocols have been developed for the detection of bacterial pathogens, including L. monocytogenes (21, 41), S. enterica (24), and S. aureus (22). Promising results obtained by this technique have also been generated with DNA extracts derived from organic waste samples (M. Lebuhn, M. Effenberger, A. Gronauer, P. A. Wilderer, and S. Wuertz, submitted for publication). Although both initial and current costs are considerably higher for real-time PCR than for conventional PCR, it might be practicable and cost efficient in standard testing laboratories when taking labor and time into account.
In conclusion, testing of nonamended samples revealed that reliable, highly sensitive detection of Salmonella spp. in organic waste samples could be performed after enrichment in peptone water at 37°C for 20 h and subsequent enrichment in Rappaport Vassiliadis medium at 43°C for 24 h prior to extraction of nucleic acids, PCR, and agarose gel electrophoresis. Those samples that tested positive by the classical detection procedure as specified in the German Biowaste Ordinance (16) could also be detected by the PCR method after two-step enrichment.
Based on this observation, it is not necessary to base a detection system on extraction of mRNA, since after two-step enrichment, it is highly unlikely that any DNA derived from dead cells is present. This is confirmed by the fact that there were no false-positive results derived from DNA of dead cells in unseeded samples, when DNA-based PCR was performed following an enrichment step. Likewise, results obtained with the other bacteria tested indicated that selective enrichment prior to nucleic acid extraction and PCR was necessary to achieve highly sensitive detection in a complex, environmental sample.
Although the procedure developed still requires selective cultivation of cells, the isolation and further differentiation of isolated colonies can be omitted, thus shortening the classical procedure. Hence, the chosen method is a compromise between the conventional enrichment technique and modern molecular biological diagnostics. In this way, the benefits of both the classical procedure (such as high sensitivity in the presence of a large number of nontarget cells and inhibitory substances) and molecular techniques (such as high efficiency) can be taken advantage of.
Acknowledgments
We thank E. Terplan for assistance in laboratory analysis and F. Tidden for invaluable advice in developing facility sampling protocols and many discussions on hygienic aspects of anaerobic digestion.
This work was funded by the Bavarian Ministry for State Development and Environmental Affairs (BAYFORREST grant F145 to S.W.).
REFERENCES
- 1.Al-Soud, W. A., and P. Rådstrõm. 1998. Capacity of nine thermostable DNA polymerases to mediate DNA amplification in the presence of PCR-inhibiting samples. Appl. Environ. Microbiol. 64:3748-3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. 1984. Compendium of methods for the microbiological examinations of foods, 2nd ed. American Public Health Association, Washington, D.C.
- 3.Baird-Parker, A. C. 1962. An improved diagnostic and selective medium for isolating coagulase positive staphylococci. J. Appl. Bacteriol. 25:12-19. [Google Scholar]
- 4.Balaban, N., and A. Rasooly. 2000. Staphylococcal enterotoxins. Int. J. Food Microbiol. 61:1-10. [DOI] [PubMed] [Google Scholar]
- 5.Brakstad, O. G., K. Aasbakk, and J. A. Maeland. 1992. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 30:1654-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bubert, A., S. Köhler, and W. Goebel. 1992. The homologous and heterologous regions within the iap gene allow genus- and species-specific identification of Listeria spp. by polymerase chain reaction. Appl. Environ. Microbiol. 58:2625-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burtscher, C., P. A. Fall, P. A. Wilderer, and S. Wuertz. 1999. Detection of Salmonella spp. and Listeria monocytogenes in suspended organic waste by nucleic acid extraction and PCR. Appl. Environ. Microbiol. 65:2235-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakraborty, T., M. Leimeister-Wächter, E. Domann, M. Hartl, W. Goebel, T. Nichterlein, and S. Notermans. 1992. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 174:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandler, D. P. 1998. Redefining relativity: quantitative PCR at low template concentrations for industrial and environmental microbiology. J. Ind. Microbiol. 21:128-140. [Google Scholar]
- 10.Cooray, K. J., T. Nishibori, H. Xiong, T. Matsuyama, M. Fujita, and M. Mitsuyama. 1994. Detection of multiple virulence-associated genes of Listeria monocytogenes by PCR in artificially contaminated milk samples. Appl. Environ. Microbiol. 60:3023-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curtis, G. D. W., R. G. Mitchell, A. F. King, and E. J. Griffin. 1989. A selective differential medium for the isolation of Listeria monocytogenes. Lett. Appl. Microbiol. 8:95-98. [Google Scholar]
- 12.D′Aoust, J. 1989. Salmonella, p. 327-445. In M. P. Doyle (ed.), Foodborne bacterial pathogens. Marcel Dekker, Inc., New York, N.Y.
- 13.Dunn, C., and W. J. Martin. 1971. Comparison of media for isolation of salmonellae and shigellae from fecal specimens. Appl. Microbiol. 22:17-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitter, S., M. Heuzenroeder, and C. J. Thomas. 1992. A combined PCR and selective enrichment method for rapid detection of Listeria monocytogenes. J. Appl. Bacteriol. 73:53-59. [DOI] [PubMed] [Google Scholar]
- 16.German Biowaste Ordinance. 1998. (Bioabfallverordnung) Verordnung über die Verwertung von Bioabfällen auf landwirtschaftlich, forstwirtschaftlich und gärtnerisch genutzten Böden, (BioAbfV) vom 21. September 1998, BGBl. I 1998 S. 2955. (In German.)
- 17.Giesendorf, B. A. J., W. G. V. Quint, M. H. C. Henkens, H. Stegeman, F. A. Huf, and H. G. M. Niesters. 1992. Rapid and sensitive detection of Campylobacter spp. in chicken products by using the polymerase chain reaction. Appl. Environ. Microbiol. 58:3804-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giolitti, G., and C. Cantoni. 1966. A medium for the isolation of staphylococci from foodstuffs. J. Appl. Bacteriol. 29:395-398. [DOI] [PubMed] [Google Scholar]
- 19.Gouws, P. A., M. Visser, and V. S. Brozel. 1998. A polymerase chain reaction procedure for the detection of Salmonella spp. within 24 hours. J. Food. Prot. 61:1039-1042. [DOI] [PubMed] [Google Scholar]
- 20.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 21.Hein, I., K. Klein, A. Lehner, A. Bubert, E. Brandl, and M. Wagner. 2001. Detection and quantification of the iap gene of Listeria monocytogenes and Listeria innocua by a new real-time quantitative PCR assay. Res. Microbiol. 152:37-46. [DOI] [PubMed] [Google Scholar]
- 22.Hein, I., A. Lehner, P. Rieck, K. Klein, E. Brandl, and M. Wagner. 2001. Comparison of different approaches to quantify Staphylococcus aureus cells by real-time quantitative PCR and application of this technique for examination of cheese. Appl. Environ. Microbiol. 67:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernandez, J., J. L. Alonso, A. Fayos, I. Amoros, and R. J. Owen. 1995. Development of a PCR assay combined with a short enrichment culture for detection of Campylobacter jejuni in estuarine surface waters. FEMS Microbiol. Lett. 127:201-206. [DOI] [PubMed] [Google Scholar]
- 24.Hoorfar, J., P. Ahrens, and P. Rådstrõm. 2000. Automated 5′ nuclease PCR assay for identification of Salmonella enterica. J. Clin. Microbiol. 38:3429-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ibrahim, A., W. Liesack, M. W. Griffiths, and R. M. Robins-Browne. 1997. Development of a highly specific assay for rapid identification of pathogenic strains of Yersinia enterocolitica based on PCR amplification of the Yersinia heat-stable enterotoxin gene (yst). J. Clin. Microbiol. 35:1636-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Josephson, K. L., C. P. Gerba, and I. L. Pepper. 1993. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl. Environ. Microbiol. 59:3513-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapperud, G. 1991. Yersinia enterocolitica in food hygiene. Int. J. Food Microbiol. 12:53-65. [DOI] [PubMed] [Google Scholar]
- 28.Klein, P. G., and V. K. Juneja. 1997. Sensitive detection of viable Listeria monocytogenes by reverse transcription PCR. Appl. Environ. Microbiol. 63:4441-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhn, M., and W. Goebel. 1989. Identification of an extracellular protein of Listeria monocytogenes possibly involved in intracellular uptake by mammalian cells. Infect. Immun. 57:55-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwaga, J., J. O. Iversen, and V. Misra. 1992. Detection of pathogenic Yersinia enterocolitica by polymerase chain reaction and digoxigenin-labeled polynucleotide probes. J. Clin. Microbiol. 30:2668-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwang, J., E. T. Littledike, and J. E. Keen. 1996. Use of the polymerase chain reaction for Salmonella detection. Lett. Appl. Microbiol. 22:46-51. [DOI] [PubMed] [Google Scholar]
- 32.Livak, K. J., S. J. Flood, J. Marmaro, W. Giusti, and K. Deetz. 1995. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 4:357-362. [DOI] [PubMed] [Google Scholar]
- 33.Loge, F. J., D. E. Thompson, and D. R. Call. 2002. PCR detection of specific pathogens in water: a risk-based analysis. Environ. Sci. Technol. 36:2754-2759. [DOI] [PubMed] [Google Scholar]
- 34.Lovell, C. R., and Y. Piceno. 1994. Purification of DNA from estuarine sediments. J. Microbiol. Methods 20:161-174. [Google Scholar]
- 35.Makino, S.-I., Y. Okada, and T. Maruyama. 1995. A new method for direct detection of Listeria monocytogenes from foods by PCR. Appl. Environ. Microbiol. 61:3745-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Flores, I., R. Cano, V. H. Bustamante, E. Calva, and J. L. Puente. 1999. The ompB operon partially determines differential expression of OmpC in Salmonella typhi and Escherichia coli. J. Bacteriol. 181:556-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McClain, D., and W. H. Lee. 1988. Development of USDA-FSIS method for isolation of Listeria monocytogenes from raw meat and poultry. J. Assoc. Off. Anal. Chem. 71:660-664. [PubMed] [Google Scholar]
- 38.McKillip, J. L., L. A. Jaykus, and M. Drake. 1999. Nucleic acid persistence in heat-killed Escherichia coli O157:H7 from contaminated skim milk. J. Food. Prot. 62:839-844. [DOI] [PubMed] [Google Scholar]
- 39.Mengaud, J., M.-F. Vicente, J. Chenevert, J. M. Pereira, C. Geoffroy, B. Gicquel-Sanzey, F. Baquero, J. C. Perez-Diaz, and P. Cossart. 1988. Expression in Escherichia coli and sequence analysis of the listeriolysin O determinant of Listeria monocytogenes. Infect. Immun. 56:766-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, V. L., J. B. Bliska, and S. Falkow. 1990. Nucleotide sequence of the Yersinia enterocolitica ail gene and characterization of the Ail protein product. J. Bacteriol. 172:1062-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nogva, H. K., K. Rudi, K. Naterstad, A. Holck, and D. Lillehaug. 2000. Application of 5′-nuclease PCR for quantitative detection of Listeria monocytogenes in pure cultures, water, skim milk, and unpasteurized whole milk. Appl. Environ. Microbiol. 66:4266-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puente, J. L., D. Juarez, M. Bobadilla, C. F. Arias, and E. Calva. 1995. The Salmonella ompC gene: structure and use as a carrier for heterologous sequences. Gene 156:1-9. [DOI] [PubMed] [Google Scholar]
- 43.Puente, J. L., V. Alvarez-Scherer, G. Gosset, and E. Calva. 1989. Comparative analysis of the Salmonella typhi and Escherichia coli ompC genes. Gene 83:197-206. [DOI] [PubMed] [Google Scholar]
- 44.Puente, J. L., A. Verdugo-Rodriguez, and E. Calva. 1991. Expression of Salmonella typhi and Escherichia coli OmpC is influenced differently by medium osmolarity: dependence on Escherichia coli OmpR. Mol. Microbiol. 5:1205-1210. [DOI] [PubMed] [Google Scholar]
- 45.Ralovich, B. 1989. Data on the enrichment and selective cultivation of listeriae. Int. J. Food Microbiol. 8:205-217. [DOI] [PubMed] [Google Scholar]
- 46.Rossen, L., K. Holmstrom, J. E. Olsen, and O. F. Rasmussen. 1991. A rapid polymerase chain reaction (PCR)-based assay for the identification of Listeria monocytogenes in food samples. Int. J. Food Microbiol. 14:145-152. [DOI] [PubMed] [Google Scholar]
- 47.Rossen, L., P. Norskov, K. Holmstrom, and O. F. Rasmussen. 1992. Inhibition of PCR by components of food samples, microbiological diagnostic assays and DNA-extraction solutions. Int. J. Food Microbiol. 17:37-45. [DOI] [PubMed] [Google Scholar]
- 48.Saiki, R. K., S. Scharf, F. Faloona, K. B. Mullis, G. T. Horn, H. A. Erlich, and N. Arnheim. 1985. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230:1350-1354. [DOI] [PubMed] [Google Scholar]
- 49.Schiemann, D. A. 1979. Synthesis of a selective agar medium for Yersinia enterocolitica. Can. J. Microbiol. 25:1298-1304. [DOI] [PubMed] [Google Scholar]
- 50.Shortle, D. 1983. A genetic system for analysis of staphylococcal nuclease. Gene 22:181-189. [DOI] [PubMed] [Google Scholar]
- 51.Smalla, K., N. Cresswell, L. C. Mendonca-Hagler, A. Wolters, and J. D. van Elsas. 1993. Rapid DNA extraction protocol from soil for polymerase chain reaction-mediated amplification. J. Appl. Bacteriol. 74:78-85. [Google Scholar]
- 52.Soumet, C., G. Ermel, P. Fach, and P. Colin. 1994. Evaluation of different DNA extraction procedures for the detection of Salmonella from chicken products by polymerase chain reaction. Lett. Appl. Microbiol. 19:294-298. [DOI] [PubMed] [Google Scholar]
- 53.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 54.Szabo, E. A., K. J. Scurrah, and J. M. Burrows. 2000. Survey for psychrotrophic bacterial pathogens in minimally processed lettuce. Lett. Appl. Microbiol. 30:456-460. [DOI] [PubMed] [Google Scholar]
- 55.Trebesius, K., D. Harmsen, A. Rakin, J. Schmelz, and J. Heesemann. 1998. Development of rRNA-targeted PCR and in situ hybridization with fluorescently labelled oligonucleotides for detection of Yersinia species. J. Clin. Microbiol. 36:2557-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsai, Y.-L., and B. H. Olson. 1991. Rapid method for direct extraction of DNA from soil and sediments. Appl. Environ. Microbiol. 57:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsai, Y. L., and B. H. Olson. 1992. Detection of low numbers of bacterial cells in soils and sediments by polymerase chain reaction. Appl. Environ. Microbiol. 58:754-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vassiliadis, F. 1984. The Rappaport-Vassiliadis (RV) enrichment medium for the isolation of salmonellae: an overview. J. Appl. Bacteriol. 54:69-76. [DOI] [PubMed] [Google Scholar]
- 59.Viswanathan, P., and R. Kaur. 2001. Prevalence and growth of pathogens on salad vegetables, fruits and sprouts. Int. J. Hyg. Environ. Health 203:205-213. [DOI] [PubMed] [Google Scholar]
- 60.Waage, A. S., T. Vardund, V. Lund, and G. Kapperud. 1999. Detection of low numbers of Salmonella in environmental water, sewage and food samples by a nested polymerase chain reaction assay. J. Appl. Microbiol. 87:418-428. [DOI] [PubMed] [Google Scholar]
- 61.Wang, R.-F., W.-W. Cao, and C. E. Cerniglia. 1996. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl. Environ. Microbiol. 62:1242-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilson, I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]