Abstract
The Agrobacterium tumefaciens C58 genome contains three putative N-acyl homoserine lactone (acyl-HSL) hydrolases, which are closely related to the lactonase AiiA of Bacillus. When expressed in Escherichia coli, two of the putative acyl-HSL hydrolases, AttM and AiiB, conferred the ability to degrade acyl-HSLs on the host. In Erwinia strain 6276, the lactonases reduced the endogenous acyl-HSL level and the bacterial virulence in planta.
N-Acyl homoserine lactones (acyl-HSLs) are diffusible signal molecules used by many gram-negative bacteria for a form of cell-to-cell communication termed quorum sensing (QS) (8). When a critical concentration of these molecules is present in the environment, i.e., when a critical cell density is reached, the acyl-HSLs bind an intracellular protein that acts as a transcriptional regulator of several genes and operons. QS regulates diverse functions, including the expression of virulence factors in several pathogenic bacteria, such as Pseudomonas aeruginosa, Agrobacterium tumefaciens, and Erwinia carotovora (Pectobacterium carotovorum) (16). Consequently, any physical or biological factors that alter the normal accumulation of acyl-HSLs may affect the virulence of such bacteria and could provide novel tools for their biological control (7). Such an approach was successfully used by Dong et al. (4): these researchers identified a lactonase enzyme in Bacillus sp. strain 204B1 that inactivates acyl-HSLs by opening the lactone ring (3). The corresponding gene, aiiA from strain 204B1 (aiiA204B1), was cloned, characterized, and expressed in plants. The resulting lactonase activity in these transgenic plants sufficed to decrease their susceptibility to infection by virulent Erwinia (4). Another gene, attM, was identified by Tn5 mutagenesis in A. tumefaciens (18). The deduced amino acid sequence of attM shows similarities with the amino acid sequences of the AiiA lactonases that are present in the Bacillus species (5, 10).
We investigated the distribution of genes homologous to the published Bacillus sp. strain 204B1 aiiA sequence (3) among the sequenced bacterial genomes available on the National Center for Biotechnology Information (NCBI) database. Using the Blastp program (http://www3.ncbi.nlm.nih.gov/BLAST/), 13 amino acid sequences deduced from open reading frames (ORFs) with higher identity scores to AiiA240B1 and belonging to eubacterial species were retained. The Bacillus sequences already identified as lactonases were excluded from this in silico search. Among these 13 ORFs, the previously identified AttM lactonase of Agrobacterium (Ag.tu.gi16119365) exhibited the best identity score (32%). In addition to this protein, two distinct putative AiiA homologues were identified in the A. tumefaciens C58 genome. To facilitate the following discussion, we termed them AiiB (Ag.tu.gi16119885) and AiiC (Ag.tu.gi17938672). While the gene encoding AttM and the aiiC locus are located on the pAt plasmid, the third locus, aiiB, lies on the pTi plasmid. The other AiiA-related ORFs (listed below) are chromosomally encoded.
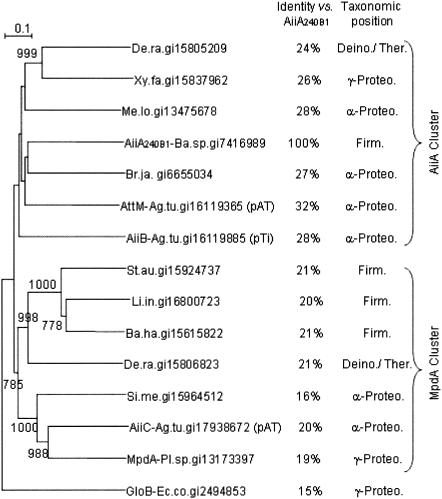
A phylogenetic analysis of these AiiA homologues showed that they fall into two clusters (Fig. 1). One cluster includes AiiA204B1, AttM, AiiB, and several AiiA homologues from the α and γ subdivisions of Proteobacteria, as well as an ORF from Deinococcus radiodurans. The second cluster encompasses the methyl parathion hydrolase from Plesiomonas sp., an ORF from each of the A. tumefaciens and Sinorhizobium meliloti genomes, and a number of putative ORFs from several gram-positive bacteria. All these ORFs matched with ORFs of one of the clusters of orthologous groups (COGs) that were defined by Tatusov et al. (14), COG0491. More than 150 prokaryotic and eukaryotic zinc metallohydrolases (or putative ORFs) are clustered in this complex phylogenetic group that encompasses a large variety of enzymes, such as glyoxalase II, class B β-lactamase, arylsulfatase, and insecticide hydrolases (2). A comparative alignment of the AiiA homologues and glyoxalase II (GloB) of Escherichia coli was performed using the ClustalW program. The results of this comparison confirmed that the most conserved regions of the AiiA homologues correspond to the characteristic domains of the Zn metallohydrolase, including the residues that are bound to the metal cations (Fig. 2). It should be emphasized that no enzymatic activity had previously been assigned to these putative ORFs, with the exceptions of the lactonase AttM (18) and the methyl parathion hydrolase MpdA in Plesiomonas sp. (19).
FIG. 1.
Phylogenetic analysis of the putative AiiA homologues. In addition to the three A. tumefaciens C58 sequences, the putative AiiA homologues were from Bacillus halodurans C-125 (Ba.ha.gi15615822), Bradyrhizobium japonicum USDA110 (Br.ja.gi6655034), Deinococcus radiodurans R1 (De.ra.gi15805209 and De.ra.gi15806823), Listeria innocua Clip11262 (Li.in.gi16800723), Mesorhizobium loti MAFF303099 (Me.lo.gi13475678), Plesiomonas sp. strain M6 (Ps.sp.gi13173397), Sinorhizobium meliloti 1021 (Si.me.gi15964512), Staphylococcus aureus Mu50 (St.au.gi15924737), and Xylella fastidiosa 9a5c (Xy.fa.gi15837962). The glyoxalase GloB (Ec.co.gi2494853) of E. coli was used as an outgroup sequence. The sequences were aligned with the ClustalW program, and the phylogenetic tree was constructed by the neighbor-joining method. Only bootstrap values greater than 750 are shown. The taxonomic position refers to the α and γ subdivisions of the Proteobacteria (α-Proteo. and γ-Proteo., respectively), the Firmicutes (Firm.), and the Deinococcus-Thermus group (Deino./Therm.).
FIG. 2.
Conserved regions in the amino acid sequences of the putative AiiA homologues. The sequences of putative AiiA homologues from Deinococcus radiodurans R1 (De.ra.gi15805209 and De.ra.gi15806823), Xylella fastidiosa 9a5c (Xy.fa.gi15837962), Mesorhizobium loti MAFF303099 (Me.lo.gi13475678), Staphylococcus aureus Mu50 (St.au.gi15924737), Listeria innocua Clip11262 (Li.in.gi16800723), Bacillus halodurans C-125 (Ba.ha.gi15615822), Sinorhizobium meliloti 1021 (Si.me.gi15964512), and Plesiomonas sp. strain M6 (Ps.sp.gi13173397) and the sequences of glyoxalase GloB (Ec.co.gi2494853) of E. coli and AiiA from Bacillus sp. strain 240B1, AiiAsoil, AttM, AiiB, and AiiC from A. tumefaciens (Ag.tu.) are shown. Each one of the five segments that are conserved among the Zn metallohydrolases (2) was present in the AiiA homologues. When more than 10 sequences (of 14) contain an identical (or a physiochemically similar residue) at a given site, the consensus residue (or the abbreviation of their physiochemical family) is reported below the alignment as follows: h for the hydrophobic residues L, I, M, and V; s for the small hydrophobic residues S, P, T, A and G; and n for the negatively charged residues and their relatives, D, E, N, and Q. The residues that are conserved among the AiiA homologues and the GloB Zn metallohydrolase are shown in bold type in the GloB sequence, and the residues interacting with the metal cations are noted by an asterisk. Numbers in parentheses indicate the number of residues omitted in the sequences.
Following this in silico analysis, we compared the hydrolytic properties of the three putative A. tumefaciens lactonases with those of AiiA of Bacillus. We focused on the A. tumefaciens lactonases for the following reasons. First, A. tumefaciens is currently the sole bacteria known to contain three ORFs closely related to the AiiA204B1 gene. Second, these proteins belong to the two different phylogenetic clusters that were defined above. Third, all these genes are located on plasmids, suggesting that they may be transferred to other bacteria. With the appropriate sets of oligonucleotides, attM (5′-GACGCAATGAAACAGAGCCG and 5′-AAGAGCGACCTGAACGAAGC), aiiB (5′-ATGCGGTTTGAGGTAGAGGC and 5′-TGAACCAGATCGCGTGACTT), and aiiC (5′-ATTTGATTGCTGGCTGAGGC and 5′-ATGGCGGAAGAAGAGGCTGT) were amplified and cloned into the pGEM cloning vector (Promega, Madison, Wis.). A Bacillus aiiA homologue was cloned using two aiiA204B1-specific primers (5′-ATGACAGTAAAGAAGCTTTATTTCG and 5′-CTATATATATTCAGGGAACACTTTAC) and DNA extracted from >1,000 bacterial colonies isolated from soil and enriched for sporeformers. To enrich for spore-forming bacteria, 20 g of soil grassland topsoil (Ordnance Survey sheet 129 grid reference 50 26) were mixed with 50 ml of 50 mM Tris (pH. 7.5), vortexed, and incubated at 80°C for 1 h to kill non-spore-forming bacteria. Samples (100 μl) of this suspension diluted 10- and 100-fold were plated onto Luria-Bertani (LB) plates and grown overnight at 28°C. Bacterial colonies were scraped directly from the plate and pooled, and genomic DNA was extracted (11). The amplified aiiA204B1 homologue was cloned into the pGEM cloning vector, sequenced, and named aiiAsoil (GenBank accession number AJ505742). Its deduced amino acid sequence and that of AiiA204B1 showed a high identity score (95%). This is consistent with a Bacillus origin for AiiAsoil, because the deduced amino acid sequences of aiiA homologues from the different Bacillus species show 90 to 96% identity with AiiA240B1 (5, 10).
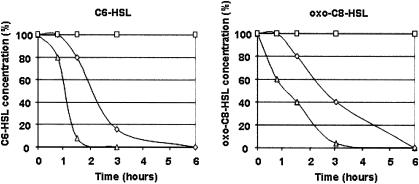
To facilitate their subsequent introduction into Erwinia (and other gram-negative bacteria), SphI-SacI fragments containing the aiiAsoil, aiiB, and aiiC genes and a NcoI-SacI fragment containing the attM gene were subcloned into the broad-host-range plasmid p6010 (9). In these p6010 derivatives, the transcription of the aiiA homologues is driven by the constitutive promoter PK. The ability of E. coli strain DH5α harboring plasmid p6010 or its derivatives pMIR101 (aiiAsoil), pMIR102 (attM), pMIR103 (aiiB), and pMIR104 (aiiC) to degrade acyl-HSLs was assayed. From a culture grown overnight in LB medium, ca. 106 bacteria were inoculated into 1 ml of fresh LB medium containing N-hexanoyl-HSL (C6-HSL), N-heptanoyl-HSL (C7-HSL), N-octanoyl-HSL (C8-HSL), N-3-oxo-hexanoyl-HSL (oxo-C6-HSL), or N-3-oxo-octanoyl-HSL (oxo-C8-HSL) at 25 μM. To prevent opening of the lactone ring under alkaline pH (17), the medium was buffered to pH 6.5 with 15 mM KH2PO4/K2HPO4. After incubation at 25°C for 24 h, 10-μl amounts of the culture medium (or appropriate dilutions for medium supplemented with oxo-C6-HSL and oxo-C8-HSL) were spotted onto thin-layer chromatography (TLC) plates for quantification as previously described (13). Samples (10 μl) of standard solutions with concentrations ranging from 25 to 1 μM (C6-HSL, C7-HSL, and C8-HSL), 250 to 10 nM (oxo-C6-HSL), and 250 to 0.4 nM (oxo-C8-HSL) and the appropriate negative controls (uninoculated medium supplemented with acyl-HSLs) were also spotted onto these TLC plates. The biosensors allowing the detection of acyl-HSLs were Chromobacterium violaceum CV026 for C6-HSL and C7-HSL (12), and A. tumefaciens NTLR4 for C8-HSL, oxo-C6-HSL, and oxo-C8-HSL (1). The experiments were done in triplicate. All the lactonases studied that belonged to the AiiA cluster conferred upon E. coli the ability to degrade acyl-HSLs. Indeed, in 24 h, more than 95% of all the acyl-HSLs disappeared in the culture media of E. coli strains harboring pMIR101, pMIR102, and pMIR103 compared with the E. coli/p6010 reference. An exception was E. coli/pMIR101, which degraded only 80% of the input oxo-C6-HSL. No disappearance of acyl-HSL was observed for E. coli carrying plasmid pMIR104, which contains the aiiC gene. We conclude that A. tumefaciens contains, in addition to attM, another locus, aiiB, that encodes a lactonase activity. The aiiB-containing DNA fragment was fully sequenced: its sequence was identical to that given by the A. tumefaciens C58 genomic databases. Following this identification step, the E. coli strains expressing the different Agrobacterium genes were compared by studying the degradation kinetics of two representative acyl-HSLs, C6-HSL and oxo-C8-HSL (A. tumefaciens produces oxo-C8-HSL). The disappearance of acyl-HSLs was monitored in bacterial cultures supplemented with acyl-HSLs (25 μM) when the cell density reached an optical density at 600 nm of 0.8. Under these experimental conditions, the AttM lactonase inactivated the input acyl-HSLs more efficiently than AiiB did (Fig. 3).
FIG. 3.
Kinetics of acyl-HSL degradation by the Agrobacterium AiiA family members. In cultures of E. coli harboring pMIR102 (attM) (▵), pMIR103 (aiiB) (⋄) and pMIR104 (aiiC) (□), the acyl-HSL concentration is expressed as a percentage of the initial concentration (25 μM) of C6-HSL and oxo-C8-HSL. The acyl-HSL degradation kinetics of control cultures of E. coli harboring p6010 and uninoculated medium supplemented with acyl-HSL were identical to those of E. coli harboring pMIR104. The experiments were done in triplicate, and the standard deviations (not shown) were always below 5%.
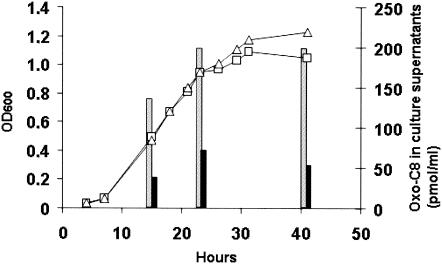
While attM is located on the catabolic plasmid pAt, aiiB lies on plasmid pTi, which harbors the acyl-HSL synthase gene, traI, as well as the functions essential for plant pathogenesis. In Agrobacterium and Bacillus, the biological role of these lactonases is still not known. It was recently hypothesized that AttM may play a role in recycling of endogenous acyl-HSLs in Agrobacterium, because AttM overexpression strongly reduced the level of oxo-C8-HSL in Agrobacterium culture medium (18). Additional evidence for this may be provided by comparing the oxo-C8-HSL levels of cultures of A. tumefaciens C58 and its derivative lacking plasmid pAt. In an A. tumefaciens C58 strain lacking pAt (15), the level of oxo-C8-HSL was always higher than in the wild type, confirming that the functions encoded by this plasmid might modulate the acyl-HSL level (Fig. 4). In addition to opine degradation (15), this would constitute additional evidence of functional cooperation between these plasmids.
FIG. 4.
Oxo-C8-HSL levels in A. tumefaciens C58 and A. tumefaciens C58 strain lacking pAt. The black and grey bars show the oxo-C8-HSL levels in the culture supernatants of the wild-type A. tumefaciens C58 strain (□) and A. tumefaciens C58 strain lacking pAt (▵), respectively. The oxo-C8-HSL concentration in wild-type strain C58 was statistically lower (P < 0.05) than in the C58 strain lacking pAt. The experiments were done in duplicate, and the standard deviations (not shown) did not exceed 10% of the mean values. OD600, optical density at 600 nm.
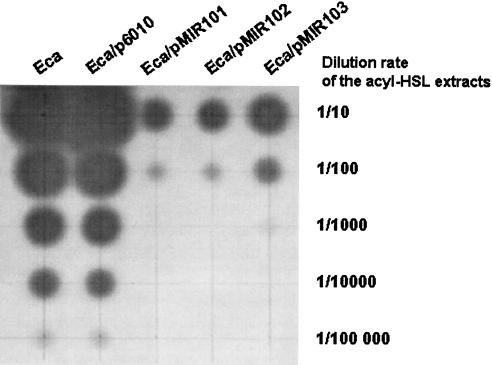
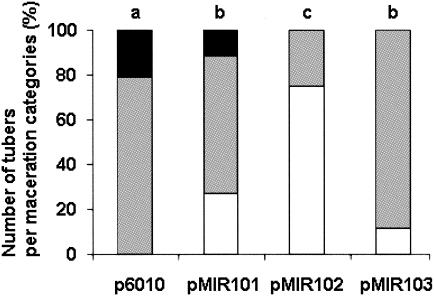
We wondered whether AiiAsoil, AttM, and AiiB might act as enzymatic antagonists to the QS-regulated virulence of pathogens, as observed for AiiA240B1 (3). To test this hypothesis, we chose the well-known phytopathogen Erwinia that expresses its virulence factors upon QS regulation (16). Plasmid p6010 and its derivatives expressing lactonases were introduced by electroporation into E. carotovora subsp. atroseptica CFBP 6276 (referred to as 6276 hereafter) (French Collection of Phytopathogenic Bacteria, Institut National de la Recherche Agronomique, Angers, France). oxo-C8-HSL was the major QS molecule produced by this E. carotovora subsp. atroseptica strain (B. Smadja and X. Latour, unpublished data). To measure the impact of lactonase expression on acyl-HSL production by strain 6276, acyl-HSLs were extracted with ethyl acetate from 6-ml samples of cultures grown overnight in LB medium at 25°C and concentrated (200 times) as described by Elasri et al. (6). These extracts were serially diluted (10−1 to 10−5), and 4-μl amounts of the diluted extracts were spotted onto a TLC plate containing the A. tumefaciens NTLR4 biosensor. The wild-type strain E. carotovora subsp. atroseptica 6276 and strain 6276 harboring p6010 produced the same amounts of acyl-HSLs. In contrast, expression of the lactonase-encoding genes reduced the concentrations of acyl-HSLs in the culture supernatant from 100-fold (pMIR103) to 1,000-fold (pMIR101 and pMIR102) (Fig. 5). Consistent with these reductions of acyl-HSL concentration in the growth medium of strain 6276 harboring the lactonase-encoding gene, an attenuated pathogenicity of strain 6276 harboring the different plasmids was observed on potatoes. For strain 6276 harboring different plasmids, ca. 20 tubers of Solanum tuberosum cv. Kaptah Vandel were inoculated at 106 CFU and incubated at 24°C under 65% humidity (Minitron; Infors HT). After 5 and 7 days of incubation, the fresh weight of macerated tissues was measured (Fig. 6). A significant decrease of maceration was observed with strain 6276 harboring all the plasmids with the lactonase-encoding genes compared to the reference strain E. carotovora subsp. atroseptica 6276/p6010. The presence of the pMIR102 plasmid that expresses AttM correlated with the highest attenuation of virulence.
FIG. 5.
Acyl-HSL levels in E. carotovora subsp. atroseptica 6276 and lactonase-expressing derivatives. The ethyl acetate extracts obtained from E. carotovora subsp. atroseptica 6276 and its lactonase-expressing derivatives were serially diluted and spotted on TLC plates and then covered by Agrobacterium NTLR4 biosensor. In the presence of the AiiA, AttM, and AiiB lactonases, the levels of acyl-HSLs in the culture supernatant decreased 100- to 1,000-fold compared to the levels of E. carotovora subsp. atroseptica 6276 and E. carotovora subsp. atroseptica 6276 harboring p6010.
FIG. 6.
Maceration assays on potatoes. In each of the tubers inoculated with E. carotovora subsp. atroseptica 6276 harboring plasmid p6010 (19 tubers), pMIR101 (26 tubers), pMIR102 (24 tubers), or pMIR103 (25 tubers), the weight of macerated tissues was measured and classified in three categories: 0 to 50 mg (white bars), 51 to 400 mg (grey bars), and more than 401 mg (black bars). The data were collected from two independent experiments. Statistically different distributions of the maceration categories (α = 0.05, χ2 test) are indicated by a different letter over the bar.
In conclusion, this work reveals that in addition to the pAt-encoded gene attM (18), A. tumefaciens harbors, on plasmid Ti, an attM-paralogous gene, aiiB, also encoding N-acyl-homoserine lactonase. Despite some data suggesting that AttM may play a role in acyl-HSL turnover, the biological and ecological functions of these two lactonases remain to be clarified through gene-by-gene mutagenesis. Finally, in addition to the aiiA genes of Bacillus (3, 4), both Agrobacterium attM and aiiB genes are suitable genes for biotechnological applications to interfere with QS-regulated virulence of pathogens, such as Erwinia.
Acknowledgments
We thank Paul Williams (Nottingham) for kindly providing acyl-HSLs.
This work was made possible by a grant (AP2001-02) from the French government (Bureau des Resources Génétiques) to D.F., by EU grant QLK3-00-31759 (Eco-safe) to R.F. and Y.D., and by a Biotechnology and Biological Sciences Research Council Sir David Phillips Fellowship awarded to R.F. S.U. was supported by a fellowship from the French government (Ministère de la Recherche et de la Technologie).
REFERENCES
- 1.Cha, C., P. Gao, Y. C. Chen, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant-Microbe Interact. 11:1119-1129. [DOI] [PubMed] [Google Scholar]
- 2.Daiyasu, H., K. Osaka, Y. Ishino, and H. Toh. 2001. Expansion of the zinc metallo-hydrolase family of the β-lactamase fold. FEBS Lett. 503:1-6. [DOI] [PubMed] [Google Scholar]
- 3.Dong, Y. H., J. L. Xu, X. Z. Li, and L. H. Zhang. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia. Proc. Natl. Acad. Sci. USA 97:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong, Y. H., L. H. Wang, H. B. Zhang, X. F. Zhang, and L. H. Zhang. 2001. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411:813-817. [DOI] [PubMed] [Google Scholar]
- 5.Dong, Y. H., A. R. Gusti, Q. Zhang, J. L. Xu, and L. H. Zhang. 2002. Identification of quorum-quenching N-acyl homoserine lactonases from Bacillus species. Appl. Environ. Microbiol. 68:1754-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elasri, M., S. Delorme, P. Lemanceau, G. Stewart, B. Laue, E. Glickmann, P. M. Oger, and Y. Dessaux. 2001. Acyl-homoserine lactone production is more common among plant-associated Pseudomonas spp. than among soilborne Pseudomonas spp. Appl. Environ. Microbiol. 67:1198-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fray, R. G. 2002. Altering plant-microbe interaction through artificially manipulating bacterial quorum sensing. Ann. Bot. 89:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heeb, S., Y. Itoh, T. Nishijyo, U. Schnider, C. Keel, J. Wade, U. Walsh, F. O'Gara, and D. Haas. 2000. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol. Plant-Microbe Interact. 13:232-237. [DOI] [PubMed] [Google Scholar]
- 10.Lee, S. J., S. Y. Park, J. J. Lee, D. Y. Yum, B. T. Koo, and J. K. Lee. 2002. Genes encoding the N-acyl homoserine lactone-degrading enzyme are widespread in many subspecies of Bacillus thuringiensis. Appl. Environ. Microbiol. 68:3919-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lippke, J. A., M. N. Strazempko, F. F. Raia, S. L. Simon, and C. K. French. 1987. Isolation of intact high-molecular-weight DNA by using guanidine isothiocyanate. Appl. Environ. Microbiol. 53:2588-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acyl homoserine lactones. Microbiology 143:3703-3711. [DOI] [PubMed] [Google Scholar]
- 13.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. Cronan, Jr., K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin layer chromatography. Proc. Natl. Acad. Sci. USA 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tatusov, R. L., D. A. Natale, I. V. Garkavtsev, T. A. Tatusova, U. T. Shankavaram, B. S. Rao, B. Kiryutin, M. Y. Galperin, N. D. Fedorova, and E. V. Koonin. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaudequin-Dransart, V., A. Petit, W. S. Chilton, and Y. Dessaux. 1998. The cryptic plasmid of Agrobacterium tumefaciens cointegrates with the Ti plasmid and cooperates for opine degradation. Mol. Plant-Microbe Interact. 11:583-591. [Google Scholar]
- 16.Whitehead, N. A., A. M. L. Barnard, H. Slater, N. J. L. Simpson, and G. P. C. Salmond. 2001. Quorum sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 17.Yates, E. A., B. Philipp, C. Buckley, S. Atkinson, S. R. Chhabra, R. E. Sockett, M. Goldner, Y. Dessaux, M. Camara, H. Smith, and P. Williams. 2002. N-Acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect. Immun. 70:5635-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang, H. B., L. H. Wang, and L. H. Zhang. 2002. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 99:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhongli, C., L. Shunpeng, and F. Guoping. 2001. Isolation of methyl parathion-degrading strain M6 and cloning of the methyl parathion hydrolase gene. Appl. Environ. Microbiol. 67:4922-4925. [DOI] [PMC free article] [PubMed] [Google Scholar]