Abstract
Cyclospora cayetanensis is a coccidian parasite that causes protracted diarrheal illness in humans. C. cayetanensis is the only species of this genus thus far associated with human illness, although Cyclospora species from other primates have been named. The current method to detect the parasite uses a nested PCR assay to amplify a 294-bp region of the small subunit rRNA gene, followed by restriction fragment length polymorphism (RFLP) or DNA sequence analysis. Since the amplicons generated from C. cayetanensis and Eimeria species are the same size, the latter step is required to distinguish between these different species. The current PCR-RFLP protocol, however, cannot distinguish between C. cayetanensis and these new isolates. The differential identification of such pathogenic and nonpathogenic parasites is essential in assessing the risks to human health from microorganisms that may be potential contaminants in food and water sources. Therefore, to expand the utility of PCR to detect and identify these parasites in a multiplex assay, a series of genus- and species-specific forward primers were designed that are able to distinguish sites of limited sequence heterogeneity in the target gene. The most effective of these unique primers were those that identified single-nucleotide polymorphisms (SNPs) at the 3′ end of the primer. Under more stringent annealing and elongation conditions, these SNP primers were able to differentiate between C. cayetanensis, nonhuman primate species of Cyclospora, and Eimeria species. As a diagnostic tool, the SNP PCR protocol described here presents a more rapid and sensitive alternative to the currently available PCR-RFLP detection method. In addition, the specificity of these diagnostic primers removes the uncertainty that can be associated with analyses of foods or environmental sources suspected of harboring potential human parasitic pathogens.
Cyclospora cayetanensis is a rapidly emerging human parasitic pathogen that causes diarrheal illness (23, 24). Whereas several types of foods have been implicated in outbreaks of human illness (1, 2, 7-10, 13, 15), evidence suggests that contaminated water sources are the primary vehicle for transmission (27-29). Molecular detection of this microorganism from environmental samples, clinical specimens, and foods has relied on a nested-PCR assay that amplifies a 294-bp fragment of the small subunit rRNA (SSU-rRNA) gene (26, 32). The same size product, however, is also amplified from species of the genus Eimeria, a closely related coccidian parasite that infects nonhuman vertebrate hosts (11). In the past, differential identification of C. cayetanensis and Eimeria species has relied on additional procedures such as either restriction fragment length polymorphism (RFLP) analysis (11) or oligonucleotide ligation assay (12). Both types of analyses are based on the limited sequence heterogeneity within the 294-bp amplified region.
Studies designed to examine the origins of C. cayetanensis and its possible host reservoirs have resulted in the discovery and description of several new primate-derived Cyclospora-like species (5, 16). Comparisons of SSU-rRNA sequence identities between the human isolate and baboon, colobus monkey, and green monkey isolates were found to be 98.9, 98.7, and 98.4% identical, respectively (5). As expected, a 294-bp PCR amplicon is produced by conventional nested-PCR method.
Although it is unclear whether the primate-derived Cyclospora species cause human illness, their presence in the environment is nonetheless a public health concern. With their potential as contaminants in water and food sources and the uncertainty of their human pathogenicity, an accurate and timely diagnosis is critical. Traditional microscopic identification that uses size and morphology to distinguish among these species is neither timely nor specific. Likewise, the lack of substantial sequence heterogeneity within the SSU-rRNA gene does not provide a means of distinguishing between human- and primate-derived Cyclospora species by the current nested-PCR-RFLP method. This becomes an increasingly important factor when the capability of a diagnostic molecular method to detect multiple infections or mixed contaminants in any sample is being considered.
Allele-specific PCR is a technique that has previously been used to differentially amplify allelic sequences that differ by at least a single base pair (6, 17-19, 30). Through the use of primers that contain defined mismatches at the 3′ end, preferential amplification of one allele relative to another can be accomplished. Using a modification of this premise, Cha et al. (3) developed a mismatch amplification mutation assay (MAMA) capable of finding a single-base-pair mutation from 100,000 copies of a wild-type allele. Manipulating conventional PCR variables of annealing and extension temperatures and time further enhanced the specificity of amplification.
In the present study, we incorporated the concepts of MAMA PCR and the conventional nested-PCR protocol for Cyclospora or Eimeria detection to develop a multiplex PCR assay to differentiate between Cyclospora and Eimeria species. By searching and comparing sequences for species-specific single-nucleotide polymorphisms (SNPs) within the SSU-rRNA gene, we designed a series of forward primers that contained the SNP at the 3′ end and would, upon specific amplification, yield a size-specific amplicon, depending upon the source of the DNA. When DNA templates were amplified under greater reaction stringencies with these SNP primers and a common reverse primer, this mismatch PCR protocol differentiated C. cayetanensis from all other species of Cyclospora and Eimeria without the need for further post-PCR analyses. In addition, we demonstrated the utility of this new PCR protocol to rapidly and successfully identify and differentiate among these related parasitic microorganisms in clinical and environmental samples.
MATERIALS AND METHODS
Parasites and parasite DNA.
C. cayetanensis oocysts were provided by J. H. Cross, Uniformed Services University of the Health Sciences, Bethesda, Md. All Eimeria oocysts were provided by R. Fayer (U.S. Department of Agriculture [BARC-East], Beltsville, Md.). The Eimeria mitis-E. mivati sample was a mixture of both species. All oocysts were stored in 2% potassium dichromate at 4°C. Oocysts of C. papionis sp.n., C. colobi sp.n., and C. cercopitheci sp.n. were provided by M. L. Eberhard (Centers for Disease Control, Atlanta, Ga.).
DNA template preparation.
DNA templates from parasite isolates (C. cayetanensis and Eimeria spp.) were prepared by applying 100 to 1,000 oocysts onto FTA filters (Fitzco, Inc., Maple Plain, Minn.) in volumes ranging from 2 to 10 μl. FTA filters were then processed as previously described and used directly as the source of DNA template for PCR (14, 21). DNA templates for C. cercopitheci, C. colobi, and C. papionis were prepared according to the protocol of Da Silva et al. (4). The approximate oocyst equivalent for these Cyclospora species was 1,000 oocysts/μl.
Conventional nested primers and PCR conditions.
Conventional PCR detection of Cyclospora species and Eimeria species was as described by Yoder et al. (32) with the nested primer pairs F1E-R2B (primary amplification) and F3E-R4B (secondary amplification) (Invitrogen Corp., Carlsbad, Calif.). For C. cayetanensis and Eimeria species, the primary amplification with primer pair F1E-R2B was performed by using prepared FTA disks as a template in a 100-μl reaction volume with HotStarTaq Master Mix (Qiagen, Valencia, Calif.) containing 200 μM concentrations each of dATP, dCTP, dGTP, and dTTP. The reaction mixture had an adjusted final concentration of 2 mM MgCl2 and 0.2 μM concentrations of primers F1E (5′-TACCCAATGAAAACAGTTT-3′) and R2B (5′-CAGGAGAAGCCAAGGTAGG-3′). For C. cercopitheci, C. colobi, and C. papionis, 2 μl of purified DNA was used as a template in a 100-μl reaction volume. All PCR amplifications were performed in a PTC-200 DNA Engine (MJ Research, Waltham, Mass.). The amplification program began with a 15 min, 95°C activation step. The cycling program consisted of 35 cycles of denaturation at 94°C for 30 s, annealing at 53°C for 30 s, and primer extension at 72°C for 90 s. A final extension cycle at 72°C for 10 min was followed by soaking at 4°C. The expected size of the primary amplicon is 636 bp. The second, nested PCR was conducted in a reaction volume of 50 μl with 1 μl of the primary amplicon as a template and primer pair F3E (5′-CCTTCCGCGCTTCGCTGCGT-3′) and R4B (5′-CGTCTTCAAACCCCCTACTG-3′). The cycling parameters were identical to first-round amplification with the exception of the annealing temperature, which was raised to 60°C. This primer pair generated a 294-bp amplicon.
RFLP analysis.
Nested PCR products obtained by using conventional primers were digested with the restriction endonuclease MnlI (11). Briefly, 20 μl of the nested reaction product was digested with 2 U of MnlI (New England Biolabs, Beverly, Mass.) in a 25-μl reaction volume for 2 h at 37°C. Digestion products were separated by gel electrophoresis with 5% NuSieve 3:1 agarose (BioWhittaker Molecular Applications, Rockland, Maine) containing ethidium bromide (0.2 μg/ml).
Species-specific primer design and multiplex PCR conditions.
Species-specific primers were designed by direct comparison of published SSU-rRNA gene sequences. These sequences were obtained from the GenBank database with the following accession numbers: C. cayetanensis, AF111183; C. cercopitheci, AF111184; C. colobi, AF111186; C. papionis, AF111187; E. acervulina, U67115; E. bovis, U77084; E. brunetti, U67116; E. maxima, U67117, E. mitis, U40262; E. mivati, U76748; E. necatrix, U67119; E. nieschulzi, U40263; E. praecox, U67120; and E. tenella, U40264. Regions of microheterogeneity within the 636-bp primary amplicon generated by using primer pair F1E and R2B (C. cayetanensis nucleotide positions 439 to 1074) were considered for nested, genus- and species-specific SNP primers (Table 1). Multiplex PCR analysis was performed by using 1 μl of the primary PCR product as a template in a 50-μl reaction volume with the same regents as described above with a 0.2 μM concentration of the common reverse primer CRP999 and 0.2 μM concentrations of each of the species-specific forward primers (Table 2). The amplification program began with an initial activation step at 95°C for 15 min, followed by a 25-cycle program of 94°C for 15 s and 66°C for 15 s. All PCR products were separated by agarose (1.5%) gel electrophoresis containing ethidium bromide (0.2 μg/ml) and then visualized on a UV transilluminator.
TABLE 1.
Sequence comparison between Cyclospora species and Eimeria species along selected regions of the SSU-rRNA gene
| Organism | GenBank accession no. | Locus sequencea (bp position)
|
||
|---|---|---|---|---|
| Nonhuman primate-derived Cyclospora spp. | C. cayetanensis | Eimeria spp. | ||
| Cyclospora spp. | ||||
| C. cayetanensis | AF111183 | CGGCCT T GCCCGT (653-665) | CGCTTC G CTGCGT (713-725) | AGGACC G AGGTAA (845-857) |
| C. cercopitheci | AF111184 | CGGCCG C GCCCGT (655-667) | CGCTTC A GTGCGT (715-727) | AGGACC G AGGTAA (848-860) |
| C. colobi | AF111186 | CGGCCG C GCCCGT (653-665) | CGCTTC A CTGCGT (713-725) | AGGACC G AGGTAA (845-857) |
| C. papionis | AF111187 | CGGCCG C GCCCGT (653-665) | CGCTTC A CTGCGT (713-725) | AGGACC G AGGTAA (845-857) |
| Baboon 22 | AF061566 | CGGCCG C GCCCGT (634-648) | CGCTTC A CTGCGT (694-706) | AGGACC G AGGTAA (827-839) |
| Baboon 34 | AF061567 | CGGCCG C GCCCGT (634-648) | CGCTTC A CTGCGT (694-706) | AGGACC G AGGTAA (827-839) |
| Baboon 40 | AF061568 | CGGCCG C GCCCGT (632-644) | CGCTTC A CTGCGT (692-704) | AGGACC G AGGTAA (825-837) |
| Eimeria spp. | ||||
| E. acervulina | U67115 | CTGCGC T GCCCGT (637-649) | CGCTTA A TTGCGT (698-710) | AGGACC A AGGTAA (831-843) |
| E. bovis | U77084 | CGGTAC C GCCCGT (633-645) | CGCTTA A TTGCTG (693-705) | AGGACC A AGGTAA (831-843) |
| E. brunetti | U67116 | CTGCGC T GCCCGT (635-647) | CTCTTA A TTGCGT (696-708) | AGGACC A AGGTAA (829-841) |
| E. maxima | U67117 | TTGCGC T GCCCGT (639-651) | GCGTTA A TTGCGT (699-711) | AGGACC A AGGTAA (833-845) |
| E. mitis | U40262 | CTGTGC T GCCCGT (636-648) | CGCTTC A CTGCGT (695-707) | AGGACC A AGGTAA (827-839) |
| E. mivati | U76748 | CTGCGC T GCCCGT (637-649) | CGCTTC A TTGCGT (698-710) | AGGACC A AGGTAA (831-843) |
| E. necatrix | U67119 | CGGCGC C GCCCGT (641-653) | CGCTTA A TTGCGT (702-714) | AGGACC A AGGTAA (835-847) |
| E. nieschulzi | U40263 | CGGTTC C GCCCGT (654-666) | CGCTTC A TTGCGT (715-727) | GGGACT A AGGTAA (848-860) |
| E. praecox | U67120 | CTGCGC T GCCCGT (636-648) | CGCTTT A TTGCGT (697-709) | AGGACC A AGGTAA (830-842) |
| E. tenella | U40264 | CGGCGT C GCCCGT (641-653) | CGCTTA A TTGCTG (702-714) | AGGACC A AGGTAA (835-847) |
A 3′-SNP used in primer design is indicated in boldface.
TABLE 2.
Mismatch PCR parameters for the differentiation of Cyclospora spp. and Eimeria spp.
| Species-specific primer | Primer specificity | Selective mismatch primer sequence | Approximate amplicon size (bp) | Nucleotide positions (GenBank no.) |
|---|---|---|---|---|
| CC719 | C. cayetanensis | 5′-GTAGCCTTCCGCGCTTCG-3′ | 298 | 711-728 (AF111183) |
| PDCL661 | C. cercopitheci, C. colobi, C. papionis | 5′-CTGTCGTGGTCATC*TGTCCGC-3′a | 361 | 641-661 (AF111184) |
| ESSP841 | Eimeria spp. | 5′-GTTCTATTTTGTTGGTTTCTAGGACCA-3′ | 174 | 815-841 (U40264) |
| CRP999 | Common reverse primer | 5′-CGTCTTCAAACCCCCTACTGTCG-3′ | 977-999 (AF111183) |
C. papionis has a substitution at the position indicated by an asterisk of G for C.
RESULTS
Identification of Cyclospora and Eimeria spp. by conventional PCR and RFLP analysis.
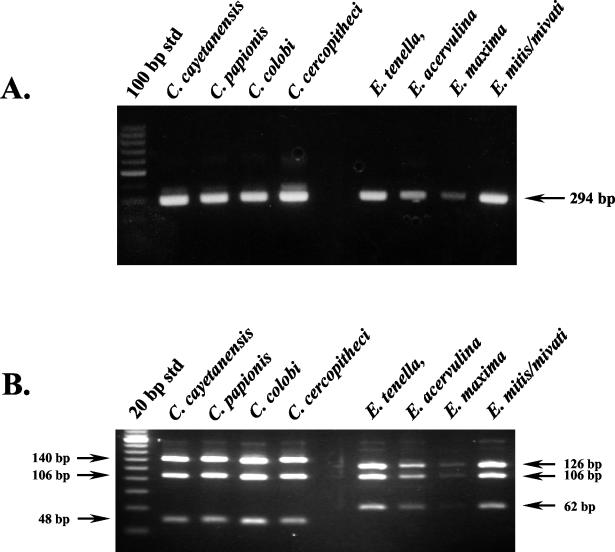
DNA templates from four Cyclospora species (C. cayetanensis, C. cercopitheci, C. colobi, and C. papionis) and four Eimeria species (E. acervulina, E. maxima, E. mivati, and E. tenella) were amplified by the conventional nested-PCR protocol with the primer pairs F1E-R2B and F3E-R4B. As expected, gel electrophoresis showed that the size of all PCR amplicons from all species were identical (Fig. 1A). Only additional analysis by RFLP differentiated between Cyclospora and Eimeria species. However, this method was unable to differentiate among the four Cyclospora species (Fig. 1B).
FIG. 1.
Molecular identification of Cyclospora and Eimeria species. (A) Conventional nested-PCR amplification. Partially purified oocysts (100 to 1,000) were spotted onto FTA filters that were used as a PCR template. For the primate-derived Cyclospora-like organisms, 2 μl of purified DNA was used as a template. A primary amplicon (636 bp [not shown]) was generated from each of these templates with the F1E-R2B primer pair, and 1 μl of this product was used in a nested amplification with the F3E-R4B primer pair to generate the 294-bp amplicon shown. (B) RFLP analysis of nested-PCR amplicons. Nested-PCR amplicons were digested with MnlI and analyzed by gel electrophoresis with 5% NuSieve 3:1 agarose containing ethidium bromide (0.2 μg/ml).
SNP primers to differentially identify C. cayetanensis.
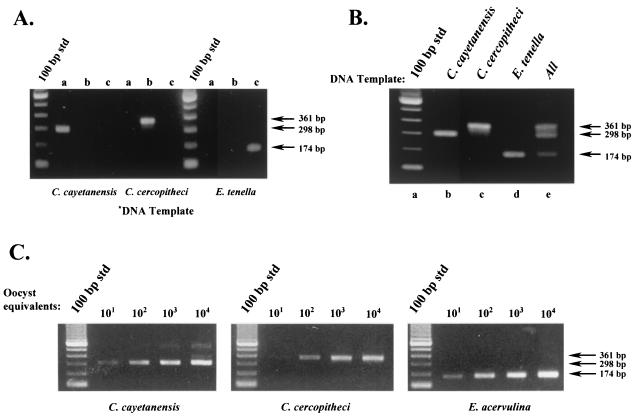
DNA templates were prepared from C. cayetanensis, C. colobi, C. cercopitheci, C. papionis, and the Eimeria species and were amplified with the conventional primer pair F1E-R2B. A multiplex reaction followed with the mismatch primers listed in Table 2. Figure 2 illustrates the selective amplification that resulted when nested reactions were performed. Individual SNP primers tested separately in the presence of primary amplicons from C. cayetanensis, C. cercopitheci, and E. tenella yielded amplicons of the expected size only when the matched DNA template and specific primer were both present (Fig. 2A). As shown, only primer pair CC719-CRP999 produced an amplicon with a C. cayetanensis-derived template. Similarly, only primer pair PDCL661-CRP999 produced an amplicon with a C. cercopitheci-derived template, and the primer pair ESSP841-CRP999 produced an amplicon with E. tenella.
FIG. 2.
Differential detection of Cyclospora and Eimeria species by using SNP primers. (A) Evaluation of SNP primer specificity. Primary PCR amplicons from C. cayetanensis, C. cercopitheci, and E. tenella were generated with the F1E-R2B primer pair. Portions (1 μl) of these products were subjected to nested-amplification reactions with their respective SNP primers as follows: lane a, SNP primer CC719; lane b, SNP primer PDCL661; and lane c, SNP primer ESSP841. (B) Multiplex PCR amplification with SNP primers to differentially detect Cyclospora and Eimeria species. Portions (1 μl) of the primary amplicons from C. cayetanensis, C. cercopitheci, and E. tenella were used as a template in a multiplex amplification reaction containing all of the SNP primers listed in Table 2 as follows: lane b, C. cayetanensis; lane c, C. cercopitheci; lane d, E. tenella; and lane e, all templates. Lane a is a standard 100-bp DNA ladder. (C) Detection sensitivity of the SNP multiplex PCR protocol. Primary PCR amplicons were prepared with the primer pair F1E-R2B. DNA templates were prepared from C. cayetanensis, C. colobi, and E. acervulina by using either intact oocysts spotted onto FTA filters (C. cayetanensis and E. acervulina) or the equivalent amount of lysate (C. colobi). Portions (3 μl) of the primary amplicons were used as a template in a multiplex amplification reaction containing all of the SNP primers as described in panel B.
As exhibited in Fig. 2B, no apparent cross-reactivity was observed as a consequence of template amplification by primers not specific for that target. Multiplex reactions that contained the complete set of SNP primers (CC719, PDCL661, ESSP841, and CRP999) and one of the individual DNA templates resulted in only one product at the predicted size for that particular organism (Fig. 2B, lanes a to d), whereas a multiplex reaction that contained DNA templates from C. cayetanensis, C. cercopitheci, and E. tenella and all three SNP primers resulted in the amplification of all three expected products (Fig. 2B, lane e).
A comparison of the sensitivity of each of the species-specific SNPs in the multiplex format (as judged by its ability to produce a detectable amplicon with decreasing amounts of their respective templates) resulted in similar levels of oocyst detection (Fig. 2C). For illustrative purposes, DNA templates from C. colobi and E. acervulina were used here to demonstrate that within a grouping (i.e., human, nonhuman primate derived, or Eimeria spp.), SNP primers would yield the expected PCR amplicon. Similar results were also obtained with C. papionis and the remaining Eimeria species listed in Table 1. As shown, the use of the SNP primers did not sacrifice assay sensitivity for the ability to differentiate Cyclospora and Eimeria spp. The SNP multiplex primer set and amplification protocol provided a detection sensitivity of the same order of magnitude as that previously demonstrated with the FTA filter-based template preparation and conventional nested-PCR primers (21). The observable difference in the lower limits of detection for C. cercopitheci compared to C. cayetanensis and E. tenella may be attributed to means by which the initial DNA template was prepared. Whereas FTA filters were used as a source of DNA template for the primary amplification of C. cayetanensis and E. tenella, nonhuman primate-derived templates (C. colobi, C. cercopitheci, and C. papionis) were prepared from oocyst lysates. More importantly, there was no indication that the presence of higher concentrations of a mismatched target gave rise to an amplicon expected for another target, i.e., high levels of a C. cayetanensis template yielding a 361- and/or 174-bp product in addition to the expected 298-bp amplicon). Multiplex reactions with one template derived from ca. 104 oocyst equivalents produced an amplicon specific only for the DNA template present.
Utility of the SNP multiplex PCR protocol with clinical and environmental samples.
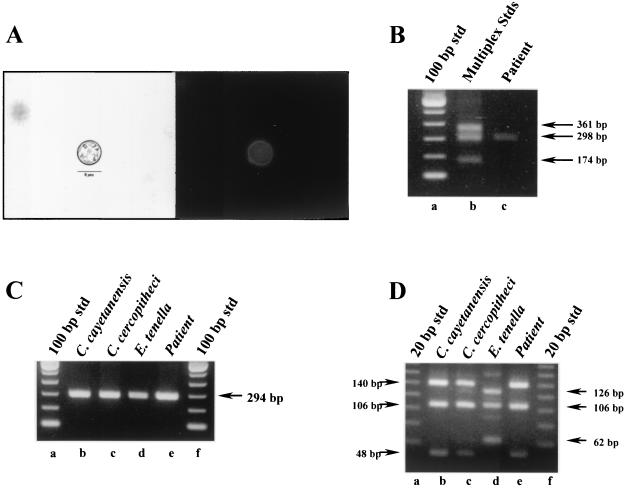
A clinical isolate and an environmental sample were used to evaluate the specificity of the SNP primers and the utility of the multiplex protocol. In the first instance, a fecal specimen was obtained from a patient that had been diagnosed with active cyclosporiasis by microscopy. Figure 3A illustrates the tentative identification of the pathogen as C. cayetanensis by its morphological and autofluorescence characteristics. This was confirmed by SNP multiplex PCR (Fig. 3B). In comparison, conventional PCR and RFLP analysis could only tentatively identify the suspected pathogen as Cyclospora but was unable to distinguish C. cayetanensis from any of the primate-derived Cyclospora species (Fig. 3C and D).
FIG. 3.
Analysis of a human fecal specimen by multiplex PCR amplification by using SNP primers to diagnose a C. cayetanensis infection. Raw fecal material (10 μl) was spotted onto an FTA filter and prepared for PCR. A primary amplicon was generated with the F1E-R2B primer pair by conventional PCR and then used in subsequent nested amplifications. (A) Microscopic identification (1000x) of C. cayetanensis by using Differential interference contrast (left panel) and UV autofluorescence (right panel) images. Magnification, ×800. (B) Multiplex PCR amplification with SNP primers. Lane a, 100-bp DNA ladder; lane b, SNP multiplex PCR amplicon standards from C. cercopitheci (360 bp), C. cayetanensis (300 bp), and E. tenella (173 bp); lane c, patient specimen. (C) Conventional nested-PCR amplification. Lanes a and f, 100-bp DNA ladder; lane b, C. cayetanensis; lane c, C. cercopitheci; lane d, E. tenella; lane e, patient specimen. (D) RFLP analysis of nested-PCR amplicons. Nested amplicons obtained in panel B were digested with MnlI. Lanes a and f, 20-bp DNA ladders; lane b, C. cayetanensis; lane c, C. cercopitheci; lane d, E. tenella; lane e, patient specimen.
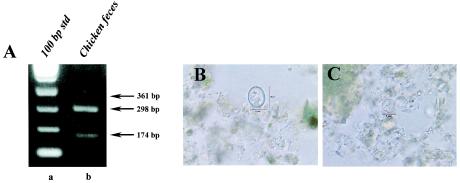
The reliability of the multiplexing capability of this protocol and the specificity of the SNP primers is shown in Fig. 4. Chicken feces obtained as a ground sample from an endemic region of Nepal was analyzed by both the SNP multiplex PCR protocol and by microscopy. PCR results indicated the presence of both C. cayetanensis and an Eimeria spp. This was later confirmed by extensive examination of the sample by microscopy (Fig. 4B and C). Although a greater proportion of the oocysts present were identified as C. cayetanensis (as judged by microscopy and as suggested by the PCR results), neither a specific count and ratio nor the species of Eimeria was determined.
FIG. 4.
Analysis of chicken feces. (A) Multiplex PCR amplification with SNP primers. (A) SNP multiplex PCR analysis of chicken feces. Raw fecal material (10 μl) was spotted onto an FTA filter and prepared for PCR. A primary amplicon was generated with the F1E-/R2B primer pair by conventional PCR and then used as DNA template in a subsequent nested amplification. Lane a, 100-bp DNA ladder; lane b, SNP multiplex PCR amplification of chicken feces. Arrows to the right of the gel indicate the amplicon sizes expected for the presence of nonhuman Cyclospora spp. (360 bp), C. cayetanensis (300 bp), and Eimeria spp. (173 bp). (B and C) Microscopic identification of an Eimeria spp. (B) and C. cayetanensis (C) by differential interference contrast. Magnification, ×800.
DISCUSSION
C. cayetanensis is an emerging human parasitic pathogen that has been associated with food-borne illness since the mid 1990s. Ongoing field studies have also detected C. cayetanensis in a number of environmental samples collected worldwide (data not shown) that extend beyond the previously identified endemic regions of Peru, Nepal, and the Caribbean islands (22, 27, 31). Since it is now regarded as a water-borne contaminant related to food-borne illness (28), this protozoan highlights the intricately intertwined relationship that exists between itself and environmental conditions, agricultural practices, food and water safety, personal hygiene, public health, and economic consequences. Research efforts to further our understanding of the environmental biology of this and other related protozoan parasites by the U.S. Food and Drug Administration, the Centers for Disease Control and Prevention, and the Environmental Protection Agency are therefore necessitated by the potential impact these pathogens may have on both public health and economic concerns.
The continued development and refinement of molecular detection methods for C. cayetanensis, such as that presented in the present study, is a major component of this effort. Here we describe the utility and selectivity of SNP primers to detect and differentiate between Cyclospora species and Eimeria species in a rapid, multiplex PCR assay. Although SSU-rRNA gene sequences among Cyclospora and Eimeria species are highly conserved, sequence comparisons within the region amplified by the primary primer pair F1E-R2B display several sites of microheterogeneity in which SNPs can be used to distinguish between these genera. Additionally, the SNP analysis also differentiated C. cayetanensis from primate-derived Cyclospora-like organisms (Table 1). From these loci, we designed a series of genus- and species-specific, nested primers (Table 2) and incorporated them into a multiplex PCR protocol. The basic design of these forward SNP primers contains a 1- to 2-bp mismatch at the penultimate and terminal bases at the 3′ end. These were subsequently used in a rapid two-step thermal cycling program that included template denaturation and a single, optimized annealing and elongation temperature (Table 2) that imposed further stringency on the amplification process. The rigor of such parameters in conjunction with the SNP primers contributed to the rapid and highly selective amplification of desired species-specific sequences. The sensitivity achieved with the SNP primers was similar to that previously reported with FTA filters for DNA template preparation and the conventional nested primers (21). In addition, we have demonstrated that this protocol can reliably detect a mixed population of pathogens in a complex sample matrix such as feces.
Several other molecular methods have also been reported that will distinguish C. cayetanensis from Eimeria species based upon sequence heterogeneity within the SSU-rRNA gene (11, 12). These methods require further analysis of the conventional nested PCR amplicon either by an RFLP or an oligoligation assay. Oliver et al. have reported another PCR method for the detection of C. cayetanensis (20). In examining sequence variability within the first internal transcribed spacer region (i.e., ITS-1) among Cyclospora species, these researchers designed unique DNA primers that detected the presence or absence of C. cayetanensis by PCR without any additional analyses. A direct comparison of the utility of these methods and that presented in the present study has not been done; however, the present protocol provides several distinct advantages for the examination of such diverse and complex matrices as foods, water sources, environmental samples, and clinical 5qolates. When coupled with the advances reported on the utility of FTA filters for the detection of human pathogens by PCR (14, 21), the use of SNP primers in a multiplex PCR protocol provides timeliness, sensitivity, and selectivity, all of which are paramount considerations in the detection of human pathogens. In a single assay, this approach also provides a means to detect and differentiate among mixed populations of similar but distinct species of microorganisms such as the closely related protozoan parasites presented here. This is an attractive component to be included in any environmental survey or epidemiological study. Likewise, this type of PCR protocol is applicable to programs designed to limit or prevent public exposure to human pathogens in fresh foods and water supplies. This is particularly important for select segments of the population that are more susceptible to infections, including the immunocompromised, the elderly, and the young. From an economic point of view, agricultural producers must receive reassurances that methods employed in routine or periodic surveillance programs are accurate and reliable. Whereas outbreaks of illness attributed to food- or water-borne contamination can be disastrous, unreliable and imprecise methodologies can have equally devastating economic consequences.
To conclude, the value of the present investigation and its results are best illustrated by what little is currently known about C. cayetanensis. Phylogenetically, Cyclospora spp. and Eimeria spp. are closely related (25, 26); however, they differ vastly in their ability to cause human disease. Whereas both genera require time and direct exposure to environmental elements to sporulate, only C. cayetanensis is known to be infectious for humans (25); at present, no information is available concerning the human pathogenicity of recently described nonhuman-primate-derived Cyclospora species. These factors all contribute, therefore, to the need for a rapid, sensitive, and accurate means not only to detect these parasites in diverse matrices (e.g., environmental samples, water sources, and fresh produce) but also to differentiate among those that pose a risk to human health. This PCR application fulfills these requirements.
REFERENCES
- 1.Centers for Disease Control. 1997. Outbreak of cyclosporiasis: Northern Virginia, Washington, D.C., Baltimore, Maryland, Metropolitan area, 1997. Morb. Moral. Wkly. Rep. 46:689-691. [PubMed] [Google Scholar]
- 2.Centers for Disease Control. 1998. Update: outbreak of cyclosporiasis—Ontario Canada, May 1998. Morb. Mortal. Wkly. Rep. 47:806-809. [PubMed] [Google Scholar]
- 3.Cha, R. S., H. Zarbl, P. Keohavong, and W. G. Thilly. 1992. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Methods Appl. 2:14-20. [DOI] [PubMed] [Google Scholar]
- 4.da Silva, A. J., F. J. Bornay-Llinares, I. N. S. Moura, S. B. Slemenda, J. L. Tuttle, and N. J. Pieniazek. 1999. Fast and reliable extraction of protozoan parasite DNA from fecal specimens. Mol. Diagn. 4:57-64. [DOI] [PubMed] [Google Scholar]
- 5.Eberhard, M. L., A. J. da Silva, B. G. Lilley, and N. J. Pieniazek. 1999. Morphological and molecular characterization of new Cyclospora species from Ethiopian monkeys: C. cercopitheci sp.n., C. colobi sp.n., and C. papionis sp.n. Emerg. Infect. Dis. 5:651-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehlen, T., and L. Dubeau. 1989. Detection of ras point mutations by polymerase chain reaction using mutation-specific, inosine-containing oligonucleotide primers. Biochem. Biophys. Res. Commun. 160:441-447. [DOI] [PubMed] [Google Scholar]
- 7.Herwaldt, B. L. 2000. Cyclospora cayetanensis: a review, focusing on the outbreaks of cyclosporiasis in the 1990's. Clin. Infect. Dis. 31:1040-1057. [DOI] [PubMed] [Google Scholar]
- 8.Herwaldt, B. L., M.-L. Ackers, et al. 1997. An outbreak in cyclosporiasis associated with imported raspberries. N. Engl. J. Med. 336:1548-1556. [DOI] [PubMed] [Google Scholar]
- 9.Herwaldt, B. L., M. J. Beach, et al. 1997. The return of Cyclospora in 1997: another outbreak of cyclosporiasis in North America associated with imported raspberries. Ann. Intern. Med. 130:210-220. [DOI] [PubMed] [Google Scholar]
- 10.Ho, A. Y., A. S. Lopez, M. G. Eberhart, R. Levenson, B. S. Finkel, A. J. da Silva, et al. 2002. Outbreak of cyclosporiasis associated with imported raspberries, Philadelphia, Pennsylvania, 2000. Emerg. Infect. Dis. 8:783-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jinneman, K. C., J. H. Wetherington, W. E. Hill, A. M. Adams, J. M. Johnson, B. J. Tenge, N.-L. Dang, R. L. Manger, and M. M. Wekell. 1998. Template preparation for PCR and RFLP of amplification products for the detection and identification of Cyclospora sp. and Eimeria sp. oocysts directly from raspberries. J. Food Prot. 61:1497-1503. [DOI] [PubMed] [Google Scholar]
- 12.Jinneman, K. C., J. H. Wetherington, W. E. Hill, C. J. Omiescinski, A. M. Adams, J. M. Johnson, B. J. Tenge, N.-L. Dang, R. L. Manger, and M. M. Wekell. 1999. An oligonucleotide-ligation assay for the differentiation between Cyclospora and Eimeria sp. polymerase chain reaction amplification products. J. Food Prot. 62:682-685. [DOI] [PubMed] [Google Scholar]
- 13.Koumans, E. H., A. D. J. Katz, J. M. Malecki, S. Kumar, S. P. Wahlquist, M. J. Arrowood, et al. 1998. An outbreak of cyclosporiasis in Florida in 1995: a harbinger of multistate outbreaks in 1996 and 1997. Am. J. Trop. Med. Hyg. 59:235-242. [DOI] [PubMed] [Google Scholar]
- 14.Lampel, K. A., P. A. Orlandi, and L. Kornegay. 2000. Improved template preparation for PCR-based assays for detection of food-borne bacterial pathogens. Appl. Environ. Microbiol. 66:4539-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez, A. S., D. R. Dodson, M. J. Arrowood, P. A. Orlandi, A. J. da Silva, J. W. Bier, et al. 2001. Outbreak of cyclosporiasis associated with basil in Missouri in 1999. Clin. Infect. Dis. 32:1010-1017. [DOI] [PubMed] [Google Scholar]
- 16.Lopez, F. A., J. Manglicmot, T. M. Schimidt, C. Yeh, H. V. Smith, and D. A. Relman. 1999. Molecular characterization of Cyclospora-like organisms from baboons. J. Infect. Dis. 179:670-676. [DOI] [PubMed] [Google Scholar]
- 17.Newton, D. R., A. Graham, L. E. Heptinstall, S. J. Powell, C. Summers, N. Kalsheker, J. C. Smith, and A. F. Markham. 1989. Analysis of any point mutation in DNA: the amplification refractory mutation system. Nucleic Acids Res. 17:2503-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nichols, W. C., J. J. Liepnieks, V. A. McKusick, and M. D. Benson. 1989. Direct sequencing of the gene for Maryland/German familial amyloidotic polyneuropathy type II and genotyping by allele-specific enzymatic amplification. Genomics 5:535-540. [DOI] [PubMed] [Google Scholar]
- 19.Okayama, H. D. T. Curiel, M. L. Brantly, M. D. Holmes, and R. G. Crystal. 1989. Rapid, nonradioactive detection of mutations in the human genome by allele-specific amplification. J. Lab. Clin. Med. 114:105-113. [PubMed] [Google Scholar]
- 20.Olivier, C., S. van de Pas, P. W. Lepp, K. Yoder, and D. A. Relman. 2001. Sequence variability in the first internal transcribed spacer region within and among Cyclospora species is consistent with polyparasitism. Int. J. Parasitol. 31:1475-1487. [DOI] [PubMed] [Google Scholar]
- 21.Orlandi, P. A., and K. A. Lampel. 2000. Extraction-free, filter-based template preparation for the rapid and sensitive PCR detection of pathogenic parasitic protozoa. J. Clin. Microbiol. 38:2271-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortega, Y., C. R. Roxas, R. H. Gilman, N. J. Miller, L. Cabrera, C. Taquiri, and C. R. Sterling. 1997. Isolation of Cryptosporidium parvum and Cyclospora cayetanensis from vegetables collected in markets of an endemic region in Peru. Am. J. Trop. Med. Hyg. 57:683-686. [DOI] [PubMed] [Google Scholar]
- 23.Ortega, Y., C. R. Sterling, R. H. Gilman, V. A. Cama, and F. Diaz. 1993. Cyclospora species-a new protozoan pathogen of humans. N. Engl. J. Med. 328:1308-1312. [DOI] [PubMed] [Google Scholar]
- 24.Ortega, Y., R. H. Gilman, and C. R. Sterling. 1994. A new coccidian parasite (Apicomplexa:Eimeriidae) from humans. J. Parasitol. 80:625-629. [PubMed] [Google Scholar]
- 25.Pieniazek, N. J., and B. L. Herwaldt. 1997. Reevaluating the molecular taxonomy: is human-associated Cyclospora a mammalian Eimeria species? Emerg. Infect. Dis. 3:381-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Relman, D. A., T. M. Schmidt, A. Gajadhar, M. Sogin, J. Cross, K. Yoder, O. Sethabutr, and P. Echeverria. 1996. Molecular phylogenetic analysis of Cyclospora, the human intestinal pathogen, suggests that it is closely related to Eimeria species. J. Infect. Dis. 173:440-445. [DOI] [PubMed] [Google Scholar]
- 27.Sherchand, J. B., J. H. Cross, M. Jimba, S. Sherchand, and M. P. Shrestha. 1999. Study of Cyclospora cayetanensis in health care facilities, sewage water and green leafy vegetables in Nepal. Southeast Asian J. Trop. Med. Public Health 30:58-63. [PubMed] [Google Scholar]
- 28.Slifko, T. R., H. V. Smith, and J. B. Rose. 2000. Emerging parasite zoonoses associated with water and food. Int. J. Parasitol. 30:1379-1393. [DOI] [PubMed] [Google Scholar]
- 29.Sturbaum, G. D., Y. R. Ortega, R. H. Gilman, C. R. Sterling, L. Cabrera, and D. Klein. 1998. Detection of Cyclospora cayetanensis in wastewater. Appl. Environ. Microbiol. 64:2284-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu, D. Y., L. Ugozzoli, B. K. Pal, and R. B. Wallace. 1989. Allele-specific enzymatic amplification of β-globin genomic DNA for diagnosis of sickle cell anemia. Proc. Natl. Acad. Sci. USA 86:2757-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wurtz, R. 1994. Cyclospora: a newly identified intestinal pathogen of humans. Clin. Infect. Dis. 18:620-623. [DOI] [PubMed] [Google Scholar]
- 32.Yoder, K. E., O. Sethabutr, and D. A. Relman. 1996. PCR-based detection of the intestinal pathogen Cyclospora, p. 169-176. In D. H. Persing (ed.), PCR protocols for emerging infectious diseases, a supplement to diagnostic molecular microbiology: principles and applications. ASM Press, Washington, D.C.