Abstract
During spring and autumn migrations, 468 fecal samples from 57 different species of migratory birds were collected in Sweden. In total, Yersinia spp. were isolated from 12.8% of collected samples. The most commonly found species was Yersinia enterocolitica, which was isolated from 5.6% of all collected samples, followed by Y. intermedia (3.8%), Y. frederiksenii (3.0%), Y. kristensenii (0.9%), Y. pseudotuberculosis (0.6%), and Y. rohdei (0.4%). The pathogenic, virF-positive Y. pseudotuberculosis strains were recovered from three thrushes. These strains belonged to the same bioserotype, 1/O:2, but had two different profiles as determined by pulsed-field gel electrophoresis with NotI and SpeI enzymes. In addition, 10 Y. enterocolitica strains, all from barnacle geese, belonged to bioserotype 3/O:3, which is associated with human disease. Two of the strains were pathogenic, carrying the virF gene on their plasmids. All pathogenic Y. pseudotuberculosis and Y. enterocolitica strains were recovered during the spring, and as the birds were caught during active migration they likely became infected at an earlier stage of the migration, thus potentially transporting these bacterial pathogens over long geographical distances.
Yersinia pseudotuberculosis and Y. enterocolitica are two species that are pathogenic for both humans and animals. While they have a nearly worldwide distribution (19), outbreaks of infection with these pathogens are rare or infrequent in most developed countries, except for Japan, where several outbreaks of Y. pseudotuberculosis infection have been reported (15, 26), and Finland, where six large outbreaks of Y. pseudotuberculosis infection have recently occurred (2). The full epidemiological pathways leading to infection in humans have not yet been elucidated, but both mammals and birds are reported to be reservoirs for these bacteria (19, 20; N. H. Harcourt-Brown, Letter, Vet. Rec. 102:315, 1978). There is a predominance of isolation of pathogenic Y. pseudotuberculosis from avian reservoirs over that of Y. enterocolitica (1, 19), and wild birds are thought to be a significant reservoir for Y. pseudotuberculosis (10, 11, 31). If pathogenic species of Yersinia are common in wild bird populations, this creates a potential for transmission of bacteria to humans and animals. Migratory birds could potentially carry bacteria along the migratory routes, allowing for dispersal of pathogenic Yersinia spp. over large geographical areas (7, 11, 14, 21). This could, in part, explain reports of Y. pseudotuberculosis and Y. enterocolitica infections on different continents (13, 16). The only existing data on the occurrence of Y. pseudotuberculosis in different migratory birds come from a survey conducted at a coastal site in Japan (13). No comparable study has been carried out to determine the occurrence of Y. pseudotuberculosis in and the risk of yersiniosis arising from wild birds in Europe.
Y. pseudotuberculosis is a relatively homogenous species that is classified into four biotypes and over 20 different serotypes, each type including pathogenic strains. In contrast, only a few combinations of Y. enterocolitica bioserotypes are classified as pathogenic to humans or animals (4). Virulence properties of Y. pseudotuberculosis and Y. enterocolitica strains can be studied with a PCR assay targeting the virulence genes on the pYV plasmid (25). DNA-based molecular methods, such as pulsed-field gel electrophoresis (PFGE), provide sensitive assays for subtyping of Y. pseudotuberculosis and Y. enterocolitica strains of the same bioserotypes (8, 17, 27). The present study investigates the occurrence of different Yersinia species in migratory birds from Sweden, providing essential data on a public health issue. We specifically look at the occurrence of pathogenic Y. pseudotuberculosis and Y. enterocolitica in the feces of birds, as well as at the virulence properties of isolated Yersinia species. PFGE combined with restriction cleavage with different enzymes was used to genotype Y. pseudotuberculosis and Y. enterocolitica isolates of the same bioserotypes, enabling us to gather information on the genetic diversity of Yersinia spp. in wild bird populations.
MATERIALS AND METHODS
Sampling.
Two locations on two different islands in the Baltic Sea in southeast Sweden were sampled: the Ottenby Bird Observatory (56°12′N, 16°24′E) on the southernmost point of Öland Island (15 March to 15 November 2000) and the coastal shore of the southern tip of Gotland Island (April 2000). Passerine birds were trapped with mistnets and Helgoland traps (5), and shorebirds were trapped with Ottenby funnel traps (5). Captured birds were banded, weighed, and measured, and their ages were determined according to differences in feather shapes and extents of wear (3, 28, 33). Three different approaches were used to obtain fecal samples, depending on the sizes of the trapped birds. Smaller birds were put one by one into a dark box with a clean sheet of paper at the bottom. Large birds, i.e., those with a body mass exceeding 250 g, were sampled by insertion of a sterile swab 1 to 2 cm into the cloaca. Samples from barnacle geese were taken by collecting fresh droppings from geese flocks grazing at the coastal meadows on Gotland Island. Sterile cotton swabs were used to collect fecal material from fresh goose droppings. Care was taken not to touch the surrounding grass. Samples were placed individually into a charcoal transport medium (Transwab; BioDisc, Solna, Sweden) and stored at 4 to 8°C for 2 to 7 days until cultivation.
Isolation and identification.
Samples were cultivated at the laboratory of the Department of Food and Environmental Hygiene at the University of Helsinki, Finland. Before direct plating, 5 ml of phosphate-buffered saline (supplemented with 1% mannitol and 0.15% bile salts according to International Organization for Standardization protocol ISO/DIS 10273) enrichment broth was added to each tube containing fecal samples. For cold enrichment, the tubes with the supplemented saline were incubated at 4°C and studied after 7, 14, and 21 days. For alkali treatment, 0.5 ml of the sample was mixed with 4.5 ml of 0.25% KOH solution for 20 s before being streaked onto CIN agar (Yersinia-selective agar base; Oxoid, Basingstoke, United Kingdom). This method was used after 7, 14, and 21 days of cold enrichment. CIN agar plates were used to isolate Yersinia, and these plates were incubated at 30°C for 18 to 20 h. To recover Y. pseudotuberculosis, a further incubation of the agar plates at 22°C for 24 h was used. Up to five whole small (diameter, <1 mm) colonies with typical a bull's eye appearance (deep red centers surrounded by outer transparent zones) on each CIN agar plate were taken and streaked onto blood agar plates for pure culture. One colony from the blood agar was inoculated onto a urea agar slant (Difco, Detroit, Mich.) and incubated for 24 h at 30°C. Isolates showing urea hydrolysis were further identified by using the Api 20E test (Biomerieux, Marcy l'Etoile, France).
Putative Y. pseudotuberculosis isolates were further biotyped according to results of reactions with raffinose, melibiose, and citrate (35). Y. enterocolitica isolates were biotyped based on assessment of the following criteria: pyrazinamidase (PYZ) and Tween-esterase activity; esculin hydrolysis; salicin, xylose, and trehalose acidification; and indole production (37). Y. pseudotuberculosis and Y. enterocolitica isolates were serotyped with slide agglutination by using a commercial serum agglutination test with O:1 to O:6 and O:3, O:5, and O:9 antisera (Denka Seiken, Tokyo, Japan), respectively.
Virulence-associated tests.
The virulence plasmids of Y. pseudotuberculosis and Y. enterocolitica isolates were studied with a PCR targeting the virF gene on the plasmids by following the modified protocol of Nakajima et al. (25). Briefly, five colonies from blood agar were suspended in 100 μl of water in one tube. DNA was extracted by boiling of the suspension for 10 min, followed by centrifugation at full speed (16,000 × g) for 3 min. Two microliters of the supernatant was used as a template in the PCR. The PCR mixture volume was 50 μl, and the mixture contained 1 U of Dynazyme DNA polymerase (Finnzymes, Espoo, Finland), 200 μM (each) deoxynucleoside triphosphates (Finnzymes), and 0.3 μM (each) primers (Pharmacia Biotech, Vantaa, Finland). PCR was performed in a 16-well PTC-150 thermal cycler (MJ Research, Watertown, Mass.). The thermal profile procedure consisted of an initial denaturation step at 94°C for 60 s followed by 29 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 60 s, and extension at 72°C for 2 min. After completion of the cyclic reactions, a final extension step at 72°C for 5 min was added. The sizes of the amplified PCR products were determined in a 0.8% agarose gel (size, 19.5 by 20 cm; 80 V for 120 min) by comparison with DNA molecular weight marker VI (Boehringer Mannheim, Mannheim, Germany). Virulence characteristics of Y. enterocolitica—i.e., reactions of PYZ, esculin, salicin (37), and calcium dependence and Congo red absorption on Congo red-magnesium oxalate agar (CR-MOX) (30)—were also tested. On CR-MOX agar, the pYV-positive Yersinia isolates produced small orange colonies (CR-MOX positive) and negative isolates produced large colorless colonies (CR-MOX negative) when incubated at 37°C for 24 h.
PFGE.
DNA extraction was performed according to the method of Niskanen et al. (27). Briefly, a single colony grown on blood agar was inoculated into 5 ml of tryptic soy broth and incubated for 18 h at room temperature. The cells from 2 ml of tryptic soy broth were washed once in 5 ml of cold PIV (10 mM Tris [pH 7.5], 1 M NaCl) and then resuspended in 750 μl of cold PIV. Next, 0.5 ml of cell suspension was mixed with 0.5 ml of 2% (wt/vol) low-melting-temperature agarose (InCert agarose; BioWhittaker Molecular Applications, Rockland, Maine) and cast in GelSyringe dispensers (New England Biolabs, Beverly, Mass.). The plugs were lysed at 37°C with shaking for 3 h in 2.5 ml of lysis solution (6 mM Tris [pH 7.5], 1 M NaCl, 100 mM EDTA [pH 7.5], 0.5% Brij-58, 0.2% sodium deoxycholate, 0.5% sodium lauroyl sarcosine, 20 μg of RNase/ml, 1 mg of lysozyme/ml) and completed with a 1-h ESP (0.5 M EDTA [pH 8.0], 10% sodium lauroyl sarcosine, 100 μg of pronase/ml) wash at 50°C. The plugs were stored at 4°C in fresh ESP solution. Before digestion, pronase was inactivated with Pefablock SC (AEBSF; Roche, Mannheim, Germany). Restriction endonuclease digestion was performed according to the manufacturer's instructions. DNA of Y. enterocolitica was digested with NotI enzyme (New England Biolabs). In addition, 10 isolates of Y. enterocolitica bioserotype 3/O:3 were also digested with SpeI enzyme (New England Biolabs). The Y. pseudotuberculosis isolates were digested both with NotI and with SpeI separately, and up to five different colonies from each positive bird were taken as replicates. The samples were electrophoresed at 12°C through a 1% (wt/vol) agarose gel (SeaKem Gold; FMC Bioproducts) in a 0.5× Tris-borate-EDTA buffer (Amresco, Solon, Ohio) at 200 V by using a Gene Navigator system (Pharmacia, Uppsala, Sweden) with a hexagonal electrode. Interpolation protocols with ranges from 1 to 18 s for 20 h for NotI and from 1 to 15 s for 18 h for SpeI were used. A midrange PFGE marker (New England Biolabs) was used for fragment size determination. The gels were stained for 30 min in 1 liter of running buffer containing 50 μl of ethidium bromide (10 mg/ml) and photographed under UV light with an Alpha Imager 2000 documentation system (Alpha Innotech, San Leandro, Calif.) by following standard procedures. For determination of NotI and SpeI profiles, the banding patterns were interpreted visually. Profiles were considered to be different when a one-band difference was observed.
RESULTS
We analyzed the occurrence of Yersinia spp. in 468 samples representing 57 different species of birds. In total, Yersinia spp. were isolated from 12.8% of all samples collected (Table 1). The most frequently encountered species was Y. enterocolitica (isolated from 5.6% of all collected fecal samples), followed by Y. intermedia (3.8%), Y. frederiksenii (3.0%), Y. kristensenii (0.9%), Y. pseudotuberculosis (0.6%), and Y. rohdei (0.4%). Most isolated strains appeared in the spring migration sample set; only five Yersinia isolates were obtained during the autumn sampling. Further, the autumn samples consisted of only nonpathogenic strains: two strains of apathogenic Y. enterocolitica biotype 1A, two strains of Y. frederiksenii, and one Y. intermedia strain.
TABLE 1.
Yersinia spp. isolated from different bird hosts
| Species | Latin name | No. tested | No. witha:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Yersinia | YP | YE | YI | YF | YK | YR | |||
| Barnacle goose | Branta leucopsis | 105 | 42 | − | 19 | 15 | 12 | 1 | 1 |
| Robin | Erithacus rubecula | 51 | − | − | − | − | − | − | − |
| Blackbird | Turdus merula | 43 | 4 | − | 2 | 1 | 1 | 2 | − |
| Redshank | Tringa totanus | 30 | 3 | − | 1 | 2 | − | − | − |
| Song thrush | Turdus philomelos | 27 | 2 | 1 | − | − | − | − | 1 |
| Dunlin | Calidris alpina | 25 | 1 | − | 1 | − | − | − | − |
| Redstart | Phoenicurus phoenicurus | 22 | 2 | − | 1 | − | 1 | − | − |
| Goldcrest | Regulus regulus | 13 | − | − | − | − | − | − | − |
| Pied flycatcher | Ficedula hypoleuca | 10 | − | − | − | − | − | − | − |
| Spotted flycatcher | Muscicapa striata | 10 | 1 | − | − | − | − | 1 | − |
| Willow warbler | Phylloscopus trochilus | 10 | − | − | − | − | − | − | − |
| Redwing | Turdus iliacus | 10 | 2 | 2 | − | − | − | − | − |
| Redpoll | Carduelis flammea | 9 | − | − | − | − | − | − | − |
| Blue tit | Parus caeruleus | 9 | − | − | − | − | − | − | − |
| Brent goose | Branta bernicla | 7 | − | − | − | − | − | − | − |
| Pied wagtail | Motacilla alba | 6 | − | − | − | − | − | − | − |
| Lesser whitethroat | Sylvia curruca | 6 | − | − | − | − | − | − | − |
| Blackcap | Sylvia atricapilla | 5 | 1 | − | 1 | − | − | − | − |
| Red-backed shrike | Lanius collurio | 4 | − | − | − | − | − | − | − |
| Garden warbler | Sylvia borin | 4 | − | − | − | − | − | − | − |
| Common whitethroat | Sylvia communis | 4 | − | − | − | − | − | − | − |
| Reed warbler | Acrocephalus scirpaceus | 3 | 1 | − | − | − | − | − | 1 |
| Rough-legged buzzard | Buteo lagopus | 3 | 1 | − | 1 | − | − | − | − |
| Jackdaw | Corvus monedula | 3 | − | − | − | − | − | − | − |
| Chiffchaff | Phylloscopus collybita | 3 | − | − | − | − | − | − | − |
| Bullfinch | Pyrrhula pyrrhula | 3 | − | − | − | − | − | − | − |
| Wren | Troglodytes troglodytes | 3 | − | − | − | − | − | − | − |
| Sparrow hawk | Accipiter nisus | 2 | − | − | − | − | − | − | − |
| Sedge warbler | Acrocephalus schoenobaenus | 2 | − | − | − | − | − | − | − |
| Tree pipit | Anthus trivialis | 2 | − | − | − | − | − | − | − |
| Siskin | Carduelis spinus | 2 | − | − | − | − | − | − | − |
| Cuckoo | Cuculus canorus | 2 | − | − | − | − | − | − | − |
| Bluethroat | Luscinia svecica | 2 | − | − | − | − | − | − | − |
| Great tit | Parus major | 2 | − | − | − | − | − | − | − |
| Starling | Sturnus vulgaris | 2 | − | − | − | − | − | − | − |
| Barred warbler | Sylvia nisoria | 2 | − | − | − | − | − | − | − |
| Fieldfare | Turdus pilaris | 2 | − | − | − | − | − | − | − |
| Marsh warbler | Acrocephalus palustris | 1 | − | − | − | − | − | − | − |
| Common sandpiper | Actites hypoleucos | 1 | − | − | − | − | − | − | − |
| Mallard | Anas platyrhynchos | 1 | − | − | − | − | − | − | − |
| Greylag goose | Anser anser | 1 | − | − | − | − | − | − | − |
| Short-eared owl | Asio flammeus | 1 | − | − | − | − | − | − | − |
| Long-eared owl | Asio otus | 1 | − | − | − | − | − | − | − |
| Canada goose | Branta canadensis | 1 | − | − | − | − | − | − | − |
| Scarlet rosefinch | Carpodacus erythrinus | 1 | − | − | − | − | − | − | − |
| Tree-creeper | Certhia familiaris | 1 | − | − | − | − | − | − | − |
| Mute swan | Cygnus olor | 1 | − | − | − | − | − | − | − |
| House martin | Delichon urbica | 1 | − | − | − | − | − | − | − |
| Great spotted woodpecker | Dendrocopus major | 1 | − | − | − | − | − | − | − |
| Ortolan bunting | Emberiza hortulana | 1 | − | − | − | − | − | − | − |
| Reed bunting | Emberiza schoeniclus | 1 | − | − | − | − | − | − | − |
| Crane | Grus grus | 1 | − | − | − | − | − | − | − |
| Icterine warbler | Hippolais icterina | 1 | − | − | − | − | − | − | − |
| Herring gull | Larus argentatus | 1 | − | − | − | − | − | − | − |
| Yellow wagtail | Motacilla flava | 1 | − | − | − | − | − | − | − |
| Grey plover | Pluvialis squatarola | 1 | − | − | − | − | − | − | − |
| Shelduck | Tadorna tadorna | 1 | − | − | − | − | − | − | − |
| Total | 57 species | 468 | 60 | 3 | 26 | 18 | 14 | 4 | 3 |
YP, Y. pseudotuberculosis; YE, Y. enterocolitica; YI, Y. intermedia; YF, Y. frederiksenii; YK, Y. kristensenii; YR, Y. rohdei; −, Yersinia not found.
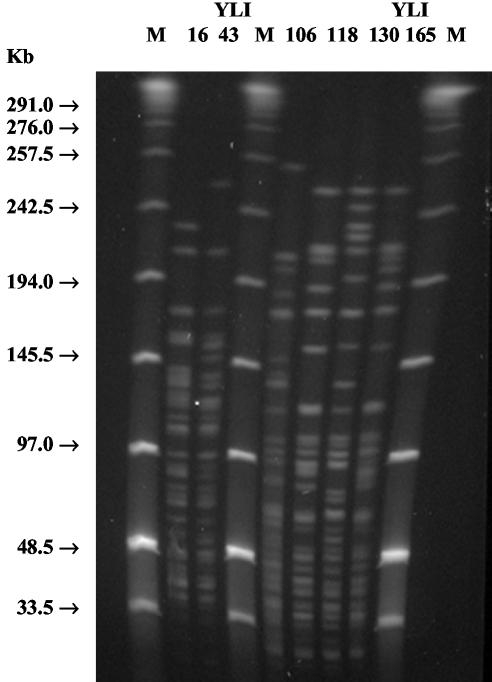
The three Y. pseudotuberculosis strains were obtained from three birds belonging to two species of thrushes, song thrush (Turdus philomelos) and redwing (Turdus iliacus), sampled during the spring migration in April. Y. pseudotuberculosis strains were of bioserotype 1/O:2, and no variation in their biochemical characteristics was present (Table 2). When virulence-associated properties of the strains were examined on CR-MOX agar plates, all of the strains showed calcium dependence and Congo red absorption. Moreover, all strains were virF-positive on PCR. The isolates of Y. pseudotuberculosis from the two bird species yielded two different PFGE profiles with both the NotI and the SpeI enzymes (Fig. 1), and this difference was consistent also when multiple Y. pseudotuberculosis colonies from the same bird were studied, meaning that the redwing and the song thrush isolates belonged to different Y. pseudotuberculosis genotypes.
TABLE 2.
Biochemical characteristics of Y. pseudotuberculosis and Y. enterocolitica strains isolated from feces of migratory birds
| Biotype/serotypea | No. of positive birds | Salicin | Esculin | PYZ | CR-MOXb | virF |
|---|---|---|---|---|---|---|
| Y. pseudotuberculosis | ||||||
| 1/O:2 | 3 | + | + | − | + | + |
| Y. enterocolitica | ||||||
| 3/O:3 | 2 | − | − | − | + | + |
| 3/O:3 | 8 | − | − | − | − | − |
| 3/NT | 9 | − | − | − | − | − |
| 1A/O:3 | 1 | + | + | + | − | − |
| 1A/NT | 3 | + | + | + | − | − |
| 1A/O:5 | 3 | + | + | + | − | − |
| NT/NT | 3 | − | − | + | − | − |
NT, nontypeable.
Isolates were tested for calcium dependence and Congo red absorption on CR-MOX agar.
FIG. 1.
Two different NotI profiles of Y. pseudotuberculosis bioserotype 1/O:2 (strains YLI16 and YLI43) and four NotI profiles of Y. enterocolitica bioserotype 3/O:3 (strains YLI106, YLI118, YLI130, and YLI165) isolated from migratory birds. M, midrange PFGE marker 1.
In bird species of which more than 10 individuals were sampled, the carriage of Y. enterocolitica varied from 0 to 18%, with the highest frequency observed in barnacle geese (Branta leucopsis) (Table 1). Of the 29 Y. enterocolitica isolates, 10 belonged to bioserotype 3/O:3, three belonged to 1A/O:5, and one belonged to 1A/O:3. The rest of the isolates did not belong to any known bioserotype combination (Table 2). Ten of the Y. enterocolitica isolates, all isolated from barnacle geese, belonged to bioserotype 3/O:3, a bioserotype associated with human disease. Two of these bioserotype 3/O:3 strains were found to be virF positive; thus, they carried the virulence plasmid. Furthermore, these isolates were PYZ negative and CR-MOX positive (Table 2). The virF-negative isolates of Y. enterocolitica bioserotype 3/O:3 were recovered from nine birds, all of which were barnacle geese. NotI enzyme digestion differentiated the 29 Y. enterocolitica isolates by producing 21 restriction patterns. The 10 isolates of bioserotype 3/O:3 corresponded to four NotI profiles (Fig. 1) and four SpeI profiles.
A total of 17 Y. pseudotuberculosis isolates were recovered after cold enrichment for 7, 14, or 21 days. However, when cold enrichment was combined with alkali treatment, 34 isolates were recovered. Four indistinguishable isolates of Y. pseudotuberculosis from one bird sample were found after direct plating. The virF-positive strains of Y. enterocolitica were isolated after a 1-week cold enrichment, followed by KOH treatment. Of the bioserotype 3/O:3 isolates, 1 and 11 were recovered after direct plating and cold enrichment, respectively.
DISCUSSION
The occurrence of Yersinia spp. among birds in this study was comparatively high, nearly 13%, and there can be no doubt that these bacteria are present in wild birds migrating through Sweden. As most of the sampled individuals were caught during active migration, the occurrence of Yersinia in samples from Öland and Gotland does not necessarily reflect the local situation in southeast Sweden. Depending on the time elapsed between colonization and excretion of the bacteria in feces and on the speed of migration of the birds, the samples may mirror the occurrence some distance away along the migratory route. As the birds sampled during the spring migration (at least all passerine birds caught at Ottenby Bird Observatory) were often caught just hours after reaching Sweden, colonized birds likely acquired the bacteria while on their wintering grounds or during the migratory journey. In our data set, there was a skewed ratio, with far more isolates obtained during spring migration than during autumn migration, and all strains of pathogenic Yersinia spp. were gathered during the spring. In interpreting results, however, one should note that the distributions of host species from which material was sampled differed between the two seasons, with some species, such as the barnacle goose, being sampled only during spring migration and others being sampled only during autumn migration, and that the number of sampled individuals was low for many species.
Most isolated Yersinia strains belonged to nonpathogenic species, namely Y. intermedia, Y. frederiksenii, Y. kristensenii, Y. rohdei, and nonpathogenic strains of Y. enterocolitica. These species have never been associated with human disease, they do not carry pYV, and the public health risk stemming from these bacteria is considered to be low. Nonpathogenic Yersinia have previously been isolated from birds, other animals, and the environment in various numbers (4, 6, 22), and differences in isolation rates can be found among different bird species in different areas (11, 23). In our study, the only frequently colonized species was the barnacle goose, of which 38% of the sampled individuals were Yersinia positive (Table 1).
Apart from the nonpathogenic strains, we also managed to isolate a few pathogenic Yersinia—three virF-positive Y. pseudotuberculosis strains and two virF-positive Y. enterocolitica strains. The strains of Y. pseudotuberculosis came from thrushes, two from song thrushes and one from a redwing, and all were retrieved during spring migration in April. Individuals of these bird species, as reflected by recoveries of banded birds trapped at Ottenby Bird Observatory, spend the nonbreeding season in Western Europe, in Italy, Spain, Portugal, France, Holland, and Belgium. All Y. pseudotuberculosis strains were of bioserotype 1/O:2. Serotype O:2 is one of the two serotypes, the other being serotype O:1, most commonly isolated from birds in Europe, the United States, and Canada. These serotypes are typically found in healthy birds but have also been reported to cause disease in both birds and humans (6, 14, 18, 34). Though serotype O:1 has been regarded as one of the most important causes of sporadic human infection with Y. pseudotuberculosis in Europe and has been associated with several outbreaks of yersiniosis, serotype O:2 also has been reported to be associated with human yersiniosis in different countries in Europe (1, 29, 34). The Y. pseudotuberculosis strains showed similar biochemical characteristics, and they all carried the virF gene on their plasmids. PFGE analysis with the NotI and SpeI enzymes divided the strains on the basis of two restriction patterns (Fig. 1). Both strains from the song thrushes clustered together and were different from the strain found in the redwing, indicating different origins of colonizing Y. pseudotuberculosis strains.
Seven different bird species, representing five different bird families (Table 1), were found to carry Y. enterocolitica. In previous studies, differences in the rates of detection of Y. pseudotuberculosis and Y. enterocolitica depending on host species have been observed, with the former more frequently isolated from passerine birds and psittaciformes while the latter has typically been isolated from birds with larger body masses (7, 14, 32). Our data were consistent with this pattern; Y. enterocolitica was most frequently isolated from barnacle geese (Table 1). Ten of the Y. enterocolitica isolates, all isolated from barnacle geese, belonged to bioserotype 3/O:3, which is associated with human disease. Two of these strains were found to be virF positive; thus, they carried the virulence plasmid. While the other eight isolates were virF negative, they should still be considered to be potentially pathogenic to humans, since accidental loss of the plasmid during isolation procedures is possible. To our knowledge, this is the first documented case of the isolation of this bioserotype from birds. Bioserotype 3/O:3 is prevalent among humans in the Far East and has been recovered from pork and chicken meat samples in Japan (12). Four of the remaining isolates of Y. enterocolitica belonged to the nonpathogenic bioserotypes 1A/O:3 and 1A/O:5, which have never been associated with human infections. The rest of the 15 Y. enterocolitica isolates did not belong to any known bioserotype combination (Table 2). Pathogenic Y. enterocolitica strains are negative for PYZ activity, esculin hydrolysis, and salicin acidification (37). The virulence characteristics differentiated virF-positive Y. enterocolitica strains from nonpathogenic Y. enterocolitica 1A strains (Table 2). Twenty-one different NotI restriction patterns were observed with Y. enterocolitica isolates. The most common NotI profile was N9, detected in samples from seven individuals—all barnacle geese. Three birds were found to carry more than one strain of Y. enterocolitica.
All samples from which Yersinia spp. were isolated in this study came from apparently healthy individuals, which has also been the case in several other studies (6, 10, 13). Y. pseudotuberculosis is a pathogen frequently observed among zoo birds and domestic fowl in connection with both outbreaks and sporadic cases of infection and is recognized to cause severe clinical symptoms and sudden death in different bird species (36, 38; Harcourt-Brown, Vet. Rec. 102:315, 1978). Perhaps wild birds are less affected by Y. pseudotuberculosis or maintain the infection at a low but latent level, developing acute clinical illness only under stressful conditions, e.g., starvation, cold temperatures, and migration.
Conventional isolation methods used in recovery of Y. pseudotuberculosis and Y. enterocolitica seem to be rather insensitive, and recovery rates are low (7, 9). Isolation of Y. pseudotuberculosis appears to be more efficient when sampling is from necropsy material instead of fecal material (7). Y. pseudotuberculosis isolates were best found after cold enrichment that was followed by KOH treatment. This has previously been shown to be the most successful method for the isolation of Y. pseudotuberculosis from clinical and environment samples with low levels of bacteria (27). The small amount of Y. pseudotuberculosis in samples makes it difficult to isolate the bacterium without enrichment since it is easily overgrown by other bacteria, including Pseudomonas, Citrobacter, Serratia, and other Yersinia spp. This could explain why Y. pseudotuberculosis was isolated only from samples in which no other Yersinia species was present. In addition, cold enrichment has been found to increase the recovery of pathogenic Y. enterocolitica strains compared with direct plating, but it also selects for nonpathogenic Y. enterocolitica strains in stool cultures (24). Our figures should thus be seen as the minimum occurrence of pathogenic Yersinia spp. in these wild birds.
Human infection with Y. pseudotuberculosis is suspected to be primarily associated with consumption of food or unchlorinated water contaminated by wild animal wastes (10). In this investigation, we show that birds caught during migration in Sweden may be infected with virF-positive Y. pseudotuberculosis and Y. enterocolitica. Birds are also commonly colonized by nonpathogenic Yersinia spp. The finding of virF-positive Y. pseudotuberculosis and Y. enterocolitica in bird feces indicates that wild birds cannot be excluded from the epidemiological discussion of human yersiniosis. However, the low isolation rate of virF-positive Y. pseudotuberculosis and Y. enterocolitica suggests that birds are unlikely to be a direct source of Yersinia infections in humans. These results provide the first knowledge on the occurrence of these largely unknown human pathogens in migratory birds in Europe and may be regarded as one step toward unraveling their epidemiology.
Acknowledgments
We thank Jaana Nieminen and Siru Salminen for their technical assistance and the ringers at Ottenby for taking the samples.
Funding for this study was provided by the Research Foundation of Orion Corporation, the Uddenberg-Nordingska Foundation, the Elis Wide Foundation, the Health Research Council of Southeast Sweden (2001-2002), and the Center for Environmental Research.
Footnotes
This is contribution no. 190 from the Ottenby Bird Observatory.
REFERENCES
- 1.Aleksic, S., J. Bockenmühl, and H. H. Wuthe. 1995. Epidemiology of Yersinia pseudotuberculosis in Germany, 1983-1993, p. 55-58. In G. Ravagnan and C. Chiesa (ed.), Contributions to microbiology and immunology, vol. 13. Yersiniosis, present and future. Karger, Basel, Switzerland. [PubMed]
- 2.Anonymous. 2000. Infectious diseases in Finland 1995-1999. KTL B4/2000. National Public Health Institute, Helsinki, Finland.
- 3.Baker, K. 1993. BTO guide 24. Identification guide to the European non-passerines. British Trust for Ornithology, Thetford, United Kingdom.
- 4.Bottone, E. J. 1997. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10:257-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bub, H. 1991. Bird trapping and bird banding. Cornell University Press, Hong Kong.
- 6.Cork, S. C., J. M. Collins-Emerson, M. R. Alley, and S. G. Fenwick. 1999. Visceral lesions caused by Yersinia pseudotuberculosis, serotype II, in different species of bird. Avian Pathol. 28:393-399. [DOI] [PubMed] [Google Scholar]
- 7.Cork, S. C., R. B. Marshall, P. Madie, and S. G. Fenwick. 1995. The role of wild birds and the environment in the epidemiology of Yersiniae in New Zealand. N. Z. Vet. J. 43:169-174. [DOI] [PubMed] [Google Scholar]
- 8.Fredriksson-Ahomaa, M., T. Autio, and H. Korkeala. 1999. Efficient subtyping of Yersinia enterocolitica bioserotype 4/O:3 with pulsed-field gel electrophoresis. Lett. Appl. Microbiol. 29:308-312. [DOI] [PubMed] [Google Scholar]
- 9.Fredriksson-Ahomaa, M., S. Hielm, and H. Korkeala. 1999. High prevalence of yadA positive Yersinia enterocolitica in pig tongues and minced meat at the retail level in Finland. J. Food Prot. 62:123-127. [DOI] [PubMed] [Google Scholar]
- 10.Fukushima, H., M. Gomyoda, and S. Kaneko. 1991. Wild animals as the source of infection with Yersinia pseudotuberculosis in Shimane Prefecture, Japan. Contrib. Microbiol. Immunol. 12:1-4. [PubMed] [Google Scholar]
- 11.Fukushima, H., and M. Gomyoda. 1991. Intestinal carriage of Yersinia pseudotuberculosis by wild birds and mammals in Japan. Appl. Environ. Microbiol. 57:1152-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukushima, H., K. Hoshina, H. Itogawa, and M. Gomyoda. 1997. Introduction into Japan of pathogenic Yersinia through imported pork, beef and fowl. Int. J. Food Microbiol. 35:205-212. [DOI] [PubMed] [Google Scholar]
- 13.Hamasaki, S., H. Hayashidani, K. Kaneko, M. Ogawa, and Y. Shigeta. 1989. A survey for Yersinia pseudotuberculosis in migratory birds in coastal Japan. J. Wildl. Dis. 25:401-403. [DOI] [PubMed] [Google Scholar]
- 14.Hubbert, W. T. 1972. Yersiniosis in mammals and birds in the United States: case reports and review. Am. J. Trop. Med. Hyg. 21:458-463. [DOI] [PubMed] [Google Scholar]
- 15.Inoue, M., H. Nakashima, T. Ishida, and M. Tsubokura. 1998. Three outbreaks of Yersinia pseudotuberculosis infection. Zentbl. Bakteriol. Mikrobiol. Hyg. 186:504-511. [PubMed] [Google Scholar]
- 16.Inoue, M., H. Nakashima, T. Mori, R. Sakazaki, K. Tamura, and M. Tsubokura. 1991. Yersinia pseudotuberculosis infection in the mountain area. Contrib. Microbiol. Immunol. 12:307-310. [PubMed] [Google Scholar]
- 17.Iteman, I., A. Guiyoule, and E. Carniel. 1996. Comparison of three molecular methods for typing and subtyping pathogenic Yersinia enterocolitica strains. J. Med. Microbiol. 45:48-56. [DOI] [PubMed] [Google Scholar]
- 18.Kageruga, P., J. Mortelmans, J. Vercruysse, and C. Beernaert-Declercq. 1976. Pseudotuberculosis in Antwerp zoo. Acta Zool. Pathol. Antverp. 66:111-120. [PubMed] [Google Scholar]
- 19.Kämpfer, P. 2000. Yersinia, p. 2342-2350. In R. K. Robinson, C. A. Batt, and P. D. Patel (ed.), Encyclopedia of food microbiology. Academic Press, San Diego, Calif.
- 20.Kaneko, K., and N. Hashimoto. 1981. Occurrence of Yersinia enterocolitica in wild animals. Appl. Environ. Microbiol. 41:635-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaneuchi, C., M. Shibata, T. Kawasaki, T. Kariu, M. Kanzaki, and T. Maruyama. 1989. Occurrence of Yersinia spp. in migratory birds, ducks, seagulls, and swallows in Japan. Jpn. J. Vet. Sci. 51:805-808. [DOI] [PubMed] [Google Scholar]
- 22.Kapperud, G., and O. Rosef. 1983. Avian wildlife reservoir of Campylobacter fetus subsp. jejuni, Yersinia spp., and Salmonella spp. in Norway. Appl. Environ. Microbiol. 46:855-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato, Y., K. Ito, Y. Kubokura, T. Maryama, K. Kaneko, and M. Ogawa. 1985. Occurrence of Yersinia enterocolitica in wild-living birds and Japanese serows. Appl. Environ. Microbiol. 49:198-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kontiainen, S., A. Sivonen, and O. V. Renkonen. 1994. Increased yields of pathogenic Yersinia enterocolitica strains by cold enrichment. Scand. J. Infect. Dis. 26:685-691. [DOI] [PubMed] [Google Scholar]
- 25.Nakajima, H., M. Inoue, T. Mori, K. I. Itoh, E. Arakawa, and H. Watanabe. 1992. Detection and identification of Yersinia pseudotuberculosis and pathogenic Yersinia enterocolitica by an improved polymerase chain reaction method. J. Clin. Microbiol. 30:2484-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakano, T., H. Kawaguchi, K. Nakao, T. Maruyama, H. Kamiya, and M. Sakurai. 1989. Two outbreaks of Yersinia pseudotuberculosis 5a infection in Japan. Scand. J. Infect. Dis. 21:175-179. [DOI] [PubMed] [Google Scholar]
- 27.Niskanen, T., M. Fredriksson-Ahomaa, and H. Korkeala. 2002. Yersinia pseudotuberculosis with limited genetic diversity is a common finding in tonsils of fattening pigs. J. Food Prot. 65:540-545. [DOI] [PubMed] [Google Scholar]
- 28.Prater, A. J., J. H. Marchant, and J. Vuorinen. 1997. BTO guide 17. Guide to the identification and ageing of holoartic waders. British Trust for Ornithology, Tring, United Kingdom.
- 29.Press, N., M. Fyfe, W. Bowie, and M. Kelly. 2001. Clinical and microbiological follow-up of an outbreak of Yersinia pseudotuberculosis serotype 1b. Scand. J. Infect. Dis. 33:523-526. [DOI] [PubMed] [Google Scholar]
- 30.Riley, G., and S. Toma. 1986. Detection of pathogenic Yersinia enterocolitica by using Congo red-magnesium oxalate agar medium. J. Clin. Microbiol. 27:213-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schiemann, D. A. 1989. Yersinia enterocolitica and Yersinia pseudotuberculosis, p. 601-672. In M. P. Doyle (ed.), Foodborne bacterial pathogens. Marcel Dekker, New York, N.Y.
- 32.Shayegani, M., W. B. Stone, I. DeForge, T. Root, L. M. Parsons, and P. Maupin. 1986. Yersinia enterocolitica and related species isolated from wildlife in New York state. Appl. Environ. Microbiol. 52:420-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svensson, L. 1992. Identification guide to European passerines. Lars Svensson, Stockholm, Sweden.
- 34.Tertti, R., K. Granfors, O. P. Lehtonen, J. Mertsola, A. L. Mäkelä, I. Välimäki, P. Hänninen, and A. Toivanen. 1984. An outbreak of Yersinia pseudotuberculosis infection. J. Infect. Dis. 149:245-250. [DOI] [PubMed] [Google Scholar]
- 35.Tsubokura, M., and S. Aleksic. 1995. A simplified antigenic scheme for serotyping of Yersinia pseudotuberculosis. Phenotypic characterization of reference strains and preparation of O and H factor sera, p. 99-105. In G. Ravagnan and C. Chiesa (ed.), Contributions to microbiology and immunology, vol. 13. Yersiniosis, present and future. Karger, Basel, Switzerland. [PubMed]
- 36.Wallner-Pendleton, E., and G. Cooper. 1983. Several outbreaks of Yersinia pseudotuberculosis in California turkey flocks. Avian Dis. 27:524-526. [PubMed] [Google Scholar]
- 37.Wauters, G., K. Kandolo, and M. Janssens. 1987. Revised biogrouping scheme of Yersinia enterocolitica. Contrib. Microbiol. Immunol. 9:14-21. [PubMed] [Google Scholar]
- 38.Welsh, R. D., R. W. Ely, and R. J. Holland. 1992. Epizootic of Yersinia pseudotuberculosis in a wildlife park. J. Am. Vet. Med. Assoc. 201:142-144. [PubMed] [Google Scholar]