Abstract
A glycoprotein (Cpgp40/15)-encoding gene of Cryptosporidium parvum was analyzed to reveal intraspecies polymorphism within C. parvum isolates. Forty-one isolates were collected from different geographical origins (Japan, Italy, and Nepal) and hosts (humans, calves, and a goat). These isolates were characterized by means of DNA sequencing, PCR-restriction fragment length polymorphism (PCR-RFLP), and RFLP-single-strand conformational polymorphism (RFLP-SSCP) analyses of the gene for Cpgp40/15. The sequence analysis indicated that there was DNA polymorphism between genotype I and II, as well as within genotype I, isolates. The DNA and amino acid sequence identities between genotypes I and II differed, depending on the isolates, ranging from 73.3 to 82.9% and 62.4 to 80.1%, respectively. Those among genotype I isolates differed, depending on the isolates, ranging from 69.0 to 85.4% and 54.8 to 79.2%, respectively. Because of the high resolution generated by PCR-RFLP and RFLP-SSCP, the isolates of genotype I could be subtyped as genotypes Ia1, Ia2, Ib, and Ie. The isolates of genotype II could be subtyped as genotypes IIa, IIb, and IIc. The isolates from calves, a goat, and one Japanese human were identified as genotype II. Within genotype II, the isolates from Japan were identified as genotype IIa, those from calves in Italy were identified as genotype IIb, and the goat isolate was identified as genotype IIc. All of the genotype I isolates were from humans. The Japanese isolate (code no. HJ3) and all of the Nepalese isolates were identified as genotypes Ia1 and Ia2, respectively. The Italian isolates were identified as genotype Ib, and the Japanese isolate (code no. HJ2) was identified as genotype Ie. Thus, the PCR-RFLP-SSCP analysis of this glycoprotein Cpgp40/15 gene generated a high resolution that has not been achieved by previous methods of genotypic differentiation of C. parvum.
Among known Cryptosporidium spp. (see the review by Xiao et al. [19]), Cryptosporidium parvum is supposedly the main species that infects humans (7). Previous studies have shown that C. parvum is mainly composed of two genotypes, I and II. The former has been found in human patients and is therefore referred to as the human type (5, 7). Genotype II was first found in cattle and is therefore referred to as the cattle type. Since then, however, this type has been found in a wide range of mammals, including humans (4, 8). Thus, both genotypes are responsible for outbreaks of human cryptosporidiosis and sporadic infections in immunocompromised individuals (6).
Despite extensive earlier studies, it is still unclear whether the pathogenesis, virulence, infectivity, or drug sensitivity of Cryptosporidium is related to some specific genotype or not. Also, genotypic information is needed to detect Cryptosporidium by means of PCR for tap water monitoring and/or to trace the infection route of the parasite.
Gene fingerprinting is a promising method with which to fulfill such requirements. Recently, one gene encoding a glycoprotein (named Cpgp40/15) of C. parvum was listed as a candidate target gene for genetic analysis of C. parvum. Sequence analysis of the Cpgp40/15 gene indicated the presence of polymorphic variants within genotype I isolates, which have been divided into five subcategories so far (2, 11). This study was undertaken to search for more advantages of the Cpgp40/15 gene for the identification and gene fingerprinting of C. parvum isolates adapting more sophisticated methods, namely, restriction fragment length polymorphism (RFLP) and single-strand conformational polymorphism (SSCP).
MATERIALS AND METHODS
Parasite isolates.
A total of 41 isolates of C. parvum were used. Six isolates were from calves in Gifu, Japan (code no. CGJ1 to CGJ6); 11 were from calves in Kobe, Japan (code no. CKJ1 to CKJ11); 1 was from a calf in Nagoya, Japan (code no. CNJ1); 8 were from calves in Italy (code no. CI1 to CI8); 1 was from a goat in Italy (code no. GI1); 3 were from humans in Japan (code no. HJ1 to HJ3); 5 were from humans in Italy (code no. HI1 to HI5); and 6 were from humans in Nepal (code no. HN1 to HN6).
All fecal samples were preserved in 2% K2Cr2O7, and oocysts were isolated by the sucrose flotation method. The samples were subjected to further purification with an immunomagnetic separation kit (Dynabeads anti-Cryptosporidium; Dynal AS, Oslo, Norway).
Template DNA for PCR was prepared as described previously (15). In brief, oocysts were frozen and thawed five times and treated at 100°C for 20 min. The samples were then digested with proteinase K at a final concentration of 200 μg/ml at 55°C for 3 h and heated at 95°C for 5 min to stop the digestion. Thus-treated samples were directly used for PCR under the conditions described in the following paragraph.
Development of PCR primer and PCR conditions.
A pair of primers (CCGTTATAGTCTCCGCTGTA and AAAGCAGAGGAACCGGCAT) were developed for amplification of the gene for Cpgy40/15 on the basis of the published sequence data (GenBank accession number AF022929) (11).
The PCR conditions used were as follows: 1 initial denaturation cycle of 94°C for 3 min; 35 cycles of 94°C for 30 s, 54°C for 30 s, and 72°C for 1 min; and 1 final extension cycle of 72°C for 10 min.
DNA sequencing.
The Cpgy40/15-encoding genes from 16 isolates (including 8 calf isolates from Japan and Italy, 1 goat isolate from Italy, and 7 human isolates from Japan, Italy, and Nepal) were sequenced. PCR products were purified with a GeneClean II Kit (Bio 101, Carlsbad, Calif.). DNA fragments were ligated into a pT7Blue T-Vector (Novagen, Inc., Madison, Wis.). The recombinant plasmids were introduced into competent cells of Escherichia coli JM109. The plasmid DNA was isolated from E. coli with a FlexiPrep Kit (Amersham Pharmacia Biotech Inc., Piscataway, N.J.).
The DNA was sequenced with a Thermo Sequenase cycle sequencing kit (USB Corporation, Cleveland, Ohio) and an automatic sequencer (LIC-4200; Aloka Co., Ltd.). The sequence data were analyzed with DNASIS software (Hitachi Software Engineering, Tokyo, Japan). Homology searching of the nucleotide and protein sequence database was carried out with the BLAST program at the National Center for Biotechnology Information (Bethesda, Md.). Pairwise sequence alignments and protein identity determinations were performed with ClustalW1.81 and PHYLIP DNADIST software.
RFLP and SSCP.
PCR-RFLP analysis was performed as previously described (14, 16). PCR products were purified by ethanol precipitation and then digested with the appropriate restriction endonucleases (AluI and RsaI) in accordance with the manufacturer's instructions.
PCR-RFLP-SSCP is a highly sensitive method developed by Wu et al. (16). In brief, the PCR-RFLP product was mixed at a 1:1 ratio with denaturing solution (95% [vol/vol] formamide, 0.02% [wt/vol] xylene cyanol, 0.02% [wt/vol] bromophenol blue, 20 mM EDTA [pH 8.0]) and heated at 95°C for 5 min. The denatured samples were cooled in ice immediately. Seven-microliter samples were applied to the gel, and the DNA samples were run at 600 V, 50 mA, 30 W, and 10°C for 110 min. After electrophoresis, the gel was stained with a PlusOne DNA silver staining kit (Amersham Pharmacia Biotech AB, Uppsala, Sweden).
RESULTS
Analysis of the Cpgp40/15 gene.
Of the 41 isolates, 16 from different geographical origins and hosts were chosen for DNA sequencing. The lengths of the amplicons produced by the primer developed for Cpgy40/15 were variable among isolates, ranging from 883 to 961 bp, and the DNA sequences of the amplicons were also diverse, as shown in Table 1 and Fig. 1.
TABLE 1.
Sequence, PCR-RFLP, and RFLP-SSCP analyses of the Cpgp40/15 gene for genotyping of C. parvum isolates
| Sequenced isolate(s) | Host | Geographical origin | DNA size (bp) | Amino acid sequence sizea | No. of serinesb | Genotype at Cpgp40/15 locus (no. tested)c |
|---|---|---|---|---|---|---|
| CGJ2, CGJ5 | Calf | Gifu, Japan | 883 | 293 | 17 | IIa (6) |
| CKJ1, CKJ3, CKJ7 | Calf | Kobe, Japan | 883 | 293 | 17 | IIa (11) |
| CNJ1 | Calf | Nagoya, Japan | 883 | 293 | 17 | IIa (1) |
| CI2, CI8 | Calf | Italy | 877 | 291 | 15 | IIb (8) |
| GI1 | Goat | Italy | 871 | 289 | 19 | IIc (1) |
| HJ1 | Human | Japan | 883 | 293 | 17 | IIa (1) |
| HJ2 | Human | Japan | 961 | 319 | 17 | Ie (1) |
| HJ3 | Human | Japan | 934 | 310 | 17 | Ia1 (1) |
| HN4, HN6 | Human | Nepal | 904 | 300 | 12 | Ia2 (6) |
| HI1, HI2 | Human | Italy | 916 | 304 | 12 | Ib (5) |
Number of amino acid residues.
Number of serine residues in the polyserine region.
Genotypes are based on sequence, PCR-RFLP, and RFLP-SSCP analyses.
FIG. 1.
DNA sequence diversities among C. parvum isolates in the Cpgp40/15 gene. The genes from 16 isolates were sequenced and tentatively grouped into seven types that are indicated on the left as IIa, IIb, IIc, Ia1, Ia2, Ib, and Ie (for the isolate names, see Materials and Methods). Dots indicate identical base pairs, and hyphens indicate gaps. The differences between IIa and IIb and between Ia1 and Ia2 are indicated by shading. The GenBank accession numbers are AY167589 (CKJ7), AY167590 (CI2), AY167591 (GI1), AY167594 (HJ3), AY167595 (HN6), AY167596 (HI2), and AY167593 (HJ2).
The DNA sequences of isolates from calves from different geographical areas of Japan (code no. CGJ2, CGJ5, CKJ1, CKJ3, CKJ7, and CNJ1) and one from a human patient in Japan (code no. HJ1) were quite similar, with more than 99% identity, and were identified as genotype II (Fig. 1).
Isolates from Italian calves (code no. CI2 and CI8) showed more than 98% DNA sequence identity with Japanese calf isolates, but there were some differences between them. The Italian isolates were 6 bp shorter than the Japanese isolates, with several base pair differences (indicated by shading in the sequence of CKJ7 and CI2 in Fig. 1). This resulted in the loss of two serine residues (15 serine repeats) in the polyserine region (Fig. 2). Therefore, we tentatively subtyped the Japanese calf isolates as genotype IIa and the Italian calf isolates as genotype IIb.
FIG. 2.
The deduced Cpgp40/15 amino acid sequences of 16 isolates were aligned by ClustalW1.81. The seven grouped types are indicated on the left as IIa, IIb, IIc, Ia1, Ia2, Ib, and Ie. Dots indicate identical base pairs, and hyphens indicate gaps. The differences between IIa and IIb and between Ia1 and Ia2 are indicated by shading.
The DNA sequences of human isolates displayed extensive polymorphism (Fig. 1 and 2 and Table 2). The DNA sequences of the human isolates, except HJ1, gave 73.3 to 82.9% identity with genotype II. Among the human isolates, the DNA sequence identities ranged from 69.0 to 85.4%, as shown in Table 2.
TABLE 2.
Genetic diversities of DNA and amino acid sequences among various C. parvum genotypes
| Genotype | Isolate(s) | Diversity (%)a
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| IIa
|
IIb, C12 | IIc, GI1 | Ia1, HJ3 | Ia2, HN6 | Ib, HI2 | Ie, HJ2 | |||||
| CKJ7 | CNJ1 | CGJ2 | HJ1 | ||||||||
| IIa | CKJ1, -3, -7 | 00.0 | 0.59 | 0.29 | 0.59 | 1.20 | 24.4 | 27.3 | 28.2 | 32.8 | 37.4 |
| CNJ1 | 0.59 | 00.0 | 0.29 | 0.59 | 1.20 | 24.5 | 27.4 | 28.3 | 33.0 | 37.6 | |
| CGJ2, CGJ5 | 0.11 | 0.11 | 00.0 | 0.29 | 0.89 | 23.9 | 26.8 | 27.8 | 32.4 | 36.9 | |
| HJ1 | 0.23 | 0.23 | 0.11 | 00.0 | 1.20 | 24.1 | 27.3 | 28.2 | 32.8 | 37.1 | |
| IIb | CI2, CI8 | 0.57 | 0.57 | 0.46 | 0.57 | 00.0 | 25.1 | 27.3 | 28.2 | 33.3 | 37.6 |
| IIc | GI1 | 16.7 | 16.7 | 16.6 | 16.6 | 17.1 | 00.0 | 28.9 | 29.9 | 28.4 | 34.6 |
| Ia1 | HJ3 | 17.3 | 17.3 | 17.2 | 17.3 | 17.5 | 18.2 | 00.0 | 0.29 | 38.9 | 45.0 |
| Ia2 | HN4, HN6 | 18.0 | 18.0 | 17.9 | 18.0 | 18.2 | 18.6 | 0.22 | 00.0 | 38.4 | 45.2 |
| Ib | HI1, HI2 | 22.5 | 22.5 | 22.3 | 22.5 | 23.3 | 17.3 | 24.8 | 24.0 | 00.0 | 20.8 |
| Ie | HJ2 | 26.7 | 26.7 | 26.5 | 26.5 | 27.2 | 22.2 | 31.0 | 30.2 | 14.6 | 00.0 |
The lower left half is DNA diversities. The upper right half is amino acid diversities.
One isolate from Japan (code no. HJ3) and the isolates from Nepal (code no. HN4 and HN6) had sequences (Fig. 1) similar to the genotype Ia sequence reported by Strong et al. (11). Although isolate HJ3 showed 99% DNA and amino acid sequence identity with the Nepalese human isolates, there were some differences among them (indicated by shading in the DNA sequences of HJ3 and HN6 in Fig. 1). There were 17 serine repeats in the polyserine region of the Japanese human isolate (code no. HJ3), while there were 12 in that of the Nepalese human isolates (HN4 and HN6); also, isolate HJ3 had an extra amino acid sequence (GKEDG; shaded in the amino acid sequences of HJ3 and HN6 in Fig. 2). Therefore, we tentatively subtyped HJ3 as genotype Ia1 and the Nepalese human isolate as genotype Ia2.
The isolates from Italian patients (HI1 and HI2) had sequences nearly identical to the genotype Ib sequence reported by Strong et al. (11). One isolate from the Japanese patient (HJ2) had a sequence nearly identical to the genotype Ie sequence reported by Leav et al. (2).
The DNA sequence of the isolate from an Italian goat (GI1) was different from both the genotype I and II isolates. The sequence identities between the Italian goat and genotype I isolates ranged from 77.8 to 82.7%, and the identities with genotype II isolates were 82.9 to 83.4% (Table 2). On the basis of the differences, this isolate was tentatively subtyped as genotype IIc.
As summarized in Table 1, the 16 isolates were tentatively grouped into seven types, including genotype IIa (Japanese isolates CGJ2, CGJ5, CKJ1, CKJ3, CKJ7, CNJ1, and HJ1), genotype IIb (Italian calf isolates CI2 and CI8), genotype IIc (Italian goat isolate GI1), genotype Ia1 (Japanese human isolate HJ3), genotype Ia2 (Nepalese human isolates HN4 and HN6), genotype Ib (Italian human isolates HI1 and HI2), and genotype Ie (Japanese human isolate HJ2).
RFLP analysis of the Cpgp40/15 gene.
On the basis of the DNA sequence difference, two endonucleases (RsaI and AluI) were chosen for RFLP analysis, predicting four kinds of RFLP patterns. Table 3 shows the predicted restriction sites of the endonuclease in representative isolates (CKJ7, CI2, IG 1, HJ2, HJ3, HI2, and HN6).
TABLE 3.
Predicted RFLP patterns of Cpgp40/15 gene PCR products
| Isolate | Sizes of predicted bands
|
Genotype | |
|---|---|---|---|
| AluI | RsaI | ||
| CKJ7 | 36, 60, 81, 206, 242, 258 | 35, 143, 177, 199, 329 | IIa |
| CI2 | 36, 60, 81, 206, 242, 258 | 35, 143, 177, 199, 329 | IIb |
| IG1 | 36, 232, 278, 325 | 35, 81, 143, 256, 356 | IIc |
| HJ3 | 146, 201, 587 | 143, 159, 241, 391 | Ia1 |
| HN6 | 131, 186, 587 | 143, 159, 241, 361 | Ia2 |
| HI2 | 56, 75, 328, 457 | 35, 143, 159, 200, 379 | Ib |
| HJ2 | 30, 128, 143, 232, 328 | 106, 129, 134, 159, 424 | Ie |
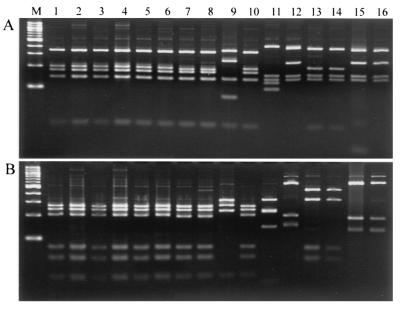
Forty-one Cryptosporidium samples were subjected to endonuclease RsaI restriction analysis. As shown in Fig. 3, calf isolates (n = 26) had exactly the same RFLP pattern, which corresponds to that of genotype II. Human isolates (n = 14) had four patterns, which correspond to those of genotypes Ia, Ib, Ie, and II. The goat isolate gave a unique pattern that is different from those of the cattle and human isolates.
FIG. 3.
PCR-RFLP analysis of the Cpgp40/15 gene by restriction with RsaI (A) and AluI (B). Japanese calf isolates (lanes 1 to 6), Italian calf isolates (lanes 7 and 8), and a Japanese human isolate HJ1 (lane 10) have the same genotype II band pattern. The Italian goat isolate (lane 9) has a unique band pattern. Japanese human isolate HJ3 (lane 12) has a band pattern similar to that of the Nepalese human isolates (lane 15 and 16), which belong to genotype Ia. The Italian human isolates (lane 13 and 14) have a unique genotype Ib band pattern. Japanese human isolate HJ2 (lane 11) has a unique genotype Ie band pattern. Lane M contains a 100-bp ladder of molecular size markers.
The same results were obtained with the AluI endonuclease, although the restriction sites of the two enzymes are different.
RFLP-SSCP analysis of the Cpgp40/15 gene.
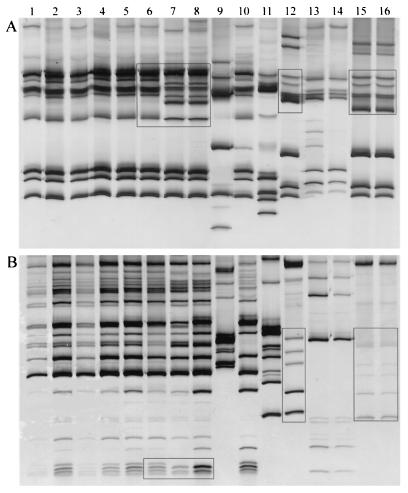
Forty-one samples of Cryptosporidium DNA were amplified by PCR, and the amplicons were restricted with endonuclease RsaI and subjected to SSCP analysis. As shown in Fig. 4, calf isolates (26 isolates) gave two SSCP patterns, corresponding to Japan (18 isolates) and Italy (8 isolates). The boxes in Fig. 4 indicate the difference between the patterns.
FIG. 4.
RFLP-SSCP analysis of the Cpgp40/15 gene. The restriction products of RsaI (A) and AluI (B) were subjected to SSCP analysis. The Japanese calf isolates (lanes 1 to 6) and Japanese human isolate HJ1 (lane 10) have a genotype IIa band pattern, and the Italian calf isolates (lanes 7 and 8) have a genotype IIb band pattern. The Italian goat isolate (lane 9) has a unique genotype IIc band pattern. Japanese human isolate HJ3 (lane 12) has a genotype Ia1 band pattern, and the Nepalese human isolates (lane 15 and 16) have a genotype Ia2 band pattern. The Italian human isolates (lane 13 and 14) have a unique genotype Ib band pattern. Japanese human isolate HJ2 (lane 11) has a unique genotype Ie band pattern. The differences between IIa and IIb and between Ia1 and Ia2 are indicated by boxes.
Fourteen human isolates were also subjected to the same analysis. Three genotypes (Ib, Ie, and II) each gave a characteristic band pattern (Fig. 4), but genotype Ia gave two band patterns; one was that of Japanese isolate HJ3, and the other was that of the six Nepalese isolates. The band pattern of the Japanese isolate (boxed in Fig. 4, lane 12) was different from those of the Nepalese isolates (boxed in Fig. 4, lanes 15 and 16). Thus, the human isolates gave five kinds of patterns.
The goat isolate gave a unique pattern that was different from those of the cattle and human isolates.
The same results were obtained by means of AluI endonuclease restriction and subsequent SSCP analysis.
DISCUSSION
In the past decade, many efforts have been made to characterize the genotype of C. parvum. DNA sequence and/or PCR-RFLP analyses of various kinds of genes, including the 18S rRNA (3, 17, 20), COWP (9, 18), HSP70 (1, 12), and TRAP (10, 13) genes, have all confirmed the genetic distinctness of the human and cattle genotypes. But the DNA sequence differences among those genes are not big enough for detection of intragenotype variation by conventional methods.
In order to identify the geographical origins of the causative agent, and to correlate its pathogenesis with specific genotypes, it is necessary to search for more diverse genes that can be used to differentiate not only between human and cattle genotypes but also within these genotypes.
In the present study, we further confirmed the usefulness of the Cpgp40/15 gene for the differentiation of isolates of C. parvum by adapting RFLP and SSCP analysis of this gene.
We have analyzed 41 samples from different geographical origins and hosts. These isolates have been genotyped on the basis of the differences at other loci, which divided these isolates into genotypes I and II (unpublished data). The DNA sequences at these loci were quite conserved, with more than 98% identity between genotypes I and II. Few variations were detected within genotypes I and II. However, the variation of the Cpgp40/15 gene sequence is quite extensive, much greater than that of any of the other Cryptosporidium loci investigated so far. The sequence differed not only between genotypes I and II (only 73.3 to 82.9% identity) but also within genotype I. The subcategories of genotype I were quite different from each other (69.8 to 85.4% identity). In fact, genotype I has been divided into five subcategories (Ia, Ib, Ic, Id, and Ie) on the basis of Cpgp40/15 sequence diversity (2, 11).
The present DNA sequence analysis can subtype the isolates analyzed into seven types. The isolates of genotype II, including the isolates from all calves, a goat, and one Japanese patient, can be subtyped into genotypes IIa, IIb, and IIc. The isolates of genotype I, including the isolates from humans in Japan, Italy, and Nepal, can be subtyped into genotypes Ia1, Ia2, Ib, and Ie. The sequences of the seven types can be distinguished by RFLP and RFLP-SSCP analyses.
One isolate from a patient in Japan and six isolates from patients in Nepal were identified as genotype I. The DNA sequences of the parasites from these two geographical regions were similar but different. This difference could be detected by the band pattern of SSCP analysis; as a consequence, the Japanese isolate was defined as Ia1 and the Nepalese isolates were defined as Ia2. The six isolates from Nepal supposedly have the same DNA sequence because the RFLP and SSCP results were identical. The isolates from Italian patients had the same sequence as the genotype Ib sequence reported by Strong et al. (11). Another Japanese patient isolate (HJ2) had a sequence similar to the genotype Ie sequence reported by Leav et al. (2).
Many gene loci, including the Cpgp40/15 locus, have been analyzed so far, but little intragenotype polymorphism has been found in genotype II. The sequence of the Cpgp40/15 gene showed that there was nearly no diversity in genotype II among the isolates from different regions of Japan. These isolates had the same length (883 bp) and the same number of serine repeats in the polyserine region. The RFLP-SSCP profiles of these isolates were similar and undistinguished, but the calf isolates from Japan and those from Italy were distinguishable by RFLP-SSCP analysis. The differences between the isolates from both geographical regions are rather small and could not be distinguished by the band pattern of RFLP analysis. However, even the small difference could be detected by RFLP-SSCP analysis, indicating the high resolution of SSCP analysis in the differentiation of even close isolates.
Our preliminary data indicate that the isolate from the Italian goat can be identified as genotype II on the basis of the RFLP results of repetitive-sequence genes (SB012, SB289, and SB281; unpublished data). The sequence of the Cpgp40/15 gene of the Italian goat, however, was very different from both the genotype I and II sequences. Therefore, the Cpgp40/15 gene analysis raises the possibility that the isolate from the Italian goat is independent from the preexisting group, genotypes I and II.
By RFLP-SSCP analysis of the Cpgp40/15 gene, all of the Japanese calf isolates had the same genotype IIa pattern. The Italian calf isolates had the genotype IIb pattern. The Nepalese human isolates had the genotype Ia1 pattern. The Italian human isolates had the genotype Ib pattern. It is likely that the genotypes are somehow related to the infection sources. Interestingly, the three Japanese patient isolates, collected at different hospitals at different times, produced three different genotypes (IIa, Ia1, and Ie). Thus, the Cpgp40/15 gene analysis by RFLP-SSCP suggests that the patients were infected by different routes.
In conclusion, PCR-RFLP and PCR-SSCP analyses of the Cpgp40/15 gene are promising methods for precise detection of genotype polymorphism among Cryptosporidium isolates that has never been performed by the analysis of other known genes. This sensitive method will provide important tools for investigation of the relationship between the genotypes and phenotypes and the molecular basis of the pathogenesis, virulence, and genetic population structure of Cryptosporidium.
Acknowledgments
This work was supported by grant 13670243 from the Ministry of Education, Science and Culture, Japan.
We thank S. Uga (Department of Medical Technology, Kobe University School of Medicine, Kobe, Japan) and E. Pozio (Laboratorio de Parasitologia, Instituto Superiore de Sanita, Rome, Italy), who kindly provided Cryptosporidium isolates.
REFERENCES
- 1.Gobet, P., and S. Toze. 2001. Sensitive genotyping of Cryptosporidium parvum by PCR-RFLP analysis of the 70-kilodalton heat shock protein (HSP70) gene. FEMS Microbiol. Lett. 200:37-41. [DOI] [PubMed] [Google Scholar]
- 2.Leav, B. A., M. R. Mackay, A. Anyanwu, R. M. O' Connor, A. M. Cevallos, G. Kindra, N. C. Rollins, M. L. Bennish, R. G. Nelson, and H. D. Ward. 2002. Analysis of sequence diversity at the highly polymorphic Cpgp40/15 locus among Cryptosporidium isolates from human immunodeficiency virus-infected children in South Africa. Infect. Immun. 70:3881-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgan, U. M., C. C. Constantine, D. A. Forbes, and R. C. A. Thompson. 1997. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. J. Parasitol. 83:825-830. [PubMed]
- 4.Morgan, U. M., D. A. Forbes, and R. C. Thompson. 1998. Molecular epidemiology of Cryptosporidium parvum. Eur. J. Protistol. 34:262-266.
- 5.Peng, M. M., L. Xiao, A. R. Freeman, M. J. Arrowood, A. A. Escalante, A. C. Weltman, C. S. Ong, W. R. Mac Kenzie, A. A. Lal, and C. B. Beard. 1997. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg. Infect. Dis. 3:567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rayer, R., U. M. Morgan, and S. L. Upton. 2000. Epidemiology of Cryptosporidium: transmission, detection and identification. Int. J. Parasitol. 30:1305-1322. [DOI] [PubMed] [Google Scholar]
- 7.Spano, F., L. Putignani, A. Crisanti, P. Sallicandro, U. M. Morgan, S. M. Le Blancq, L. Tchack, S. Tzipori, and G. Widmer. 1998. A multilocus genotypic analysis of Cryptosporidium parvum from different hosts and geographical origins. J. Clin. Microbiol. 36:3255-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spano, F., L. Putignani, S. Guida, and A. Crisanti. 1999. Cryptosporidium parvum: PCR-RFLP analysis of the TRAP-C1 (thrombosondin-related adhesive protein of Cryptosporidium-1) gene discriminates between two alleles differentially associated with parasite isolates of animal and human origin. Exp. Parasitol. 90:195-198. [DOI] [PubMed]
- 9.Spano, F., L. Putignani, J. McLauchlin, D. P. Casemore, and A. Crisanti. 1997. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol. Lett. 150:209-217. [DOI] [PubMed]
- 10.Spano, F., L. Putignani, S. Naitza, C. Puri, S. Wright, and A. Crisanti. 1998. Molecular cloning and expression analysis of a Cryptosporidium parvum gene encoding a new member of the thrombospondin family. Mol. Biochem. Parasitol. 92:147-162. [DOI] [PubMed]
- 11.Strong, W. B., J. Gut, and R. G. Nelson. 2000. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect. Immun. 68:4117-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sulaiman, I. M., U. M. Morgan, R. C. A. Thompson, A. A. Lal, and L. Xiao. 2000. Phylogenetic relationships of Cryptosporidium parasites based on the 70-kilodalton heat shock protein (HSP) gene. Appl. Environ. Microbiol. 66:2385-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sulaiman, I. M., L. Xiao, C. Yang, L. Escalante, A. Moore, C. B. Beard, M. J. Arrowood, and A. A. Lal. 1998. Differentiating human from animal isolates of Cryptosporidium parvum. Emerg. Infect. Dis. 4:681-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu, Z., I. Nagano, A. Matsuo, E. Pozio, and Y. Takahashi. 1999. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) for the identification of Trichinella isolates. Parasitology 118:211-218. [DOI] [PubMed] [Google Scholar]
- 15.Wu, Z., I. Nagano, A. Matsuo, S. Uga, I. Kimata, M. Iseki, and Y. Takahashi. 2000. Specific PCR primers for Cryptosporidium parvum with extra high sensitivity. Mol. Cell. Probes 14:33-39. [DOI] [PubMed] [Google Scholar]
- 16.Wu, Z., I. Nagano, T. Nakada, and Y. Takahashi. 1999. DNA fingerprints of Trichinella as revealed by restriction fragment length polymorphism and single-strand conformational polymorphism (RFLP-SSCP). Mol. Cell. Probes 14:291-297. [DOI] [PubMed] [Google Scholar]
- 17.Xiao, L., L. Escalante, C. Yang, I. Sulaiman, A. A. Escalante, R. J. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small subunit ribosomal RNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao, L., J. Kimor, U. M. Morgan, I. M. Sulaiman, R. C. A. Thompson, and A. A. Lal. 2000. Sequence differences in the diagnostic target region of the oocyst wall protein gene of Cryptosporidium parasites. Appl. Environ. Microbiol. 66:5499-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao, L., U. M. Morgan, R. Fayer, R. C. A. Thompsom, and A. A. Lal. 2000. Cryptosporidium systematics and implications for public health. Parasitol. Today 16:287-292. [DOI] [PubMed] [Google Scholar]
- 20.Xiao, L., U. M. Morgan, J. Limor, A. Escalante, M. Arrowood, W. Shulaw, R. C. A. Thompson, R. Fayer, and A. A. Lal. 1999. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 65:3386-3391. [DOI] [PMC free article] [PubMed]